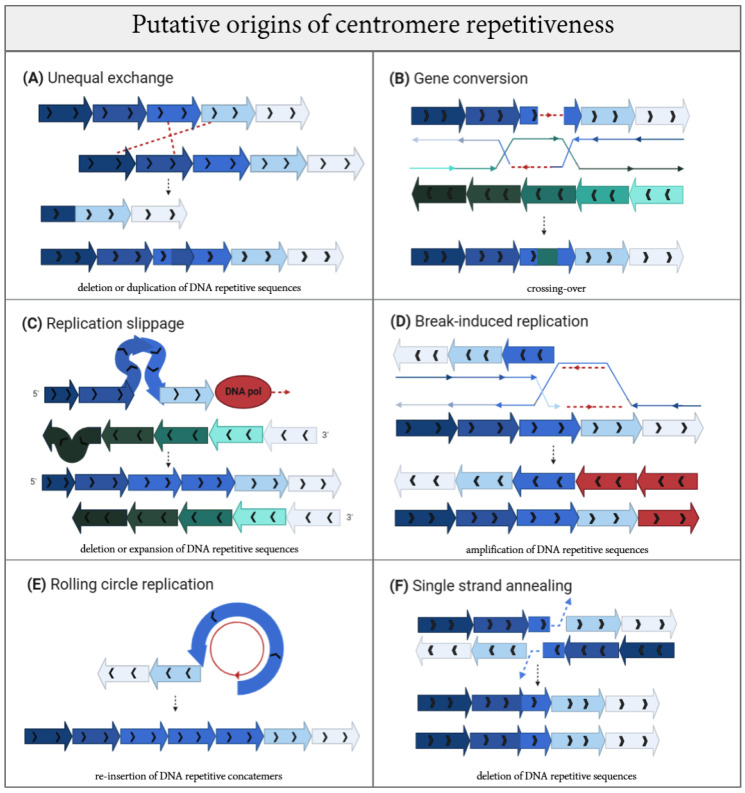
Figure 3.
Mutagenic processes that may operate at centromere sequences and have contributed to their repetitive origins. (A) Unequal exchange following recombination can cause gain or loss of tandem repeats and DNA rearrangements. (B) Gene conversion causes the unidirectional transfer of genetic information among homologous repetitive DNA sequences and can result in reciprocal or non-reciprocal exchange (the latter is depicted). (C) Replication slippage on misalignment repeated DNA strands during replication is thought to induce centromere expansion or contraction depending on whether the hairpin (depicted)/distortion is found on the newly synthesized strand (blue repeats) or the bulge (depicted)/distortion is on the template DNA (green repeats). (D) Break-induced replication (BIR) repairs one-ended double-stranded break (DSB) substrate, produced by replication fork collapse. (E) Rolling circle replication occurs when the 3′ end circularizes, and its replication produces repeated concatemers. (F) Single strand annealing (SSA) repairs DSBs through the annealing of complementary ssDNA strands succeeded by DNA tail end digestion and ligation. These repair pathways are essential for maintaining genome stability, yet when operating on repetitive sequences (especially arranged in tandem and sharing high degree of sequence homology like at the centromere), they may result in mutagenic variability as a way for ongoing DNA evolution and shaping.