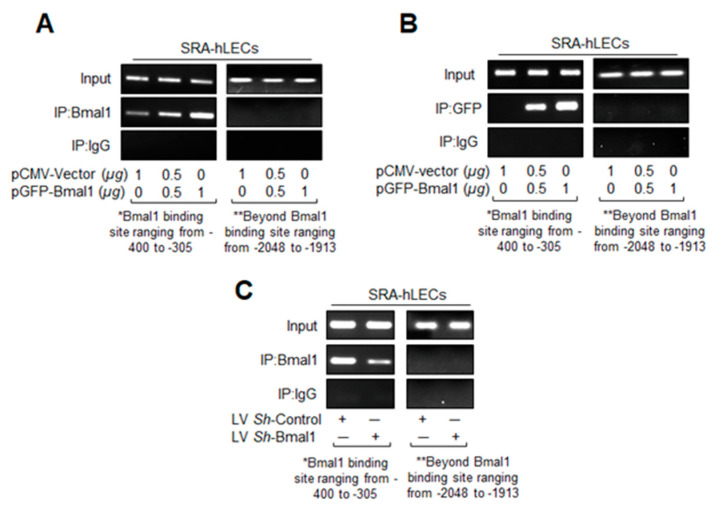
Figure 5.
In vivo DNA binding assay revealed that cellular abundance of Bmal1 influenced its binding to E-box sequences present in Prdx6 promoter. Diagrammatic sketch of the 5′-constructs of human Prdx6 gene promoter containing Bmal1-DNA binding sites and ChIP primers location and sequences used for ChIP are depicted in Figure 4A. (A,B) DNA binding experiments with LECs overexpressing Bmal1 showed its increased binding to its responsive elements in the Prdx6 gene promoter. Chromatin samples were prepared from SRA-hLECs overexpressed with pCMV-vector or pGFP-Bmal1 plasmid and were submitted to ChIP assay with ChIP grade antibodies anti-Bmal1 (A), anti-GFP (B), and anti-IgG. Immunoprecipitated DNA fragments were purified and processed for PCR analysis using primers that specifically recognize fragments of the Prdx6 promoter containing the Bmal1 binding site (*, -400 to -305) as indicated. PCR products were resolved onto an agarose gel and visualized with ethidium bromide staining. Photographs are representative of three experiments. Data revealed a significant augmentation of Bmal1 binding to E-Box in a dose-dependent manner. (C) Bmal1 knockdown SRA-hLECs displayed a significant loss in Bmal1 binding to E-Box in the Prdx6 gene promoter in vivo. Genomic DNA was cross-linked to immobilize bound protein in vivo, was sheared and immunoprecipitated with anti-Bmal1 or unrelated control IgG, and was amplified using PCR with primers specific to the region. The quality of input DNA was initially measured and equalized by optical density (O.D.) Representative photographic images of the amplified DNA band visualized with ethidium bromide staining are shown. Primers designed beyond the Bmal1 binding sites (**, -2048 to -1913) or a mock ChIP with control IgG was used as a negative control.