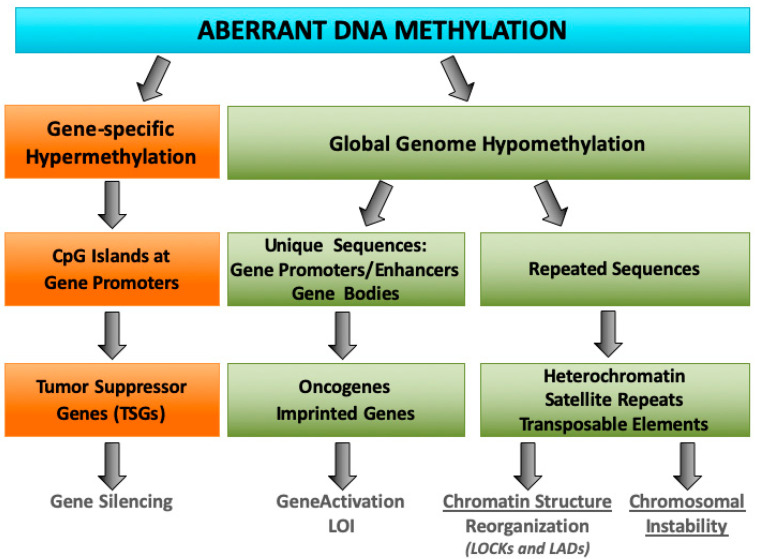
Figure 1.
Scheme of aberrant DNA methylation in cancer cells. Left (orange): Locus-specific hypermethylation of CpG island in promoter sequences leads to transcriptional inactivation of tumor suppressor genes in cancer cells. Right (green): Global cancer-associated hypomethylation affects both unique and repeated sequences. Hypomethylation of unique sequences participates to the activation of oncogenes, mediated by transcriptional enhancers and LOI of imprinted genes involved in cell growth control and tumorigenesis. Hypomethylation of tandem repeats (centromeric and juxta-centromeric satellite DNA), interspersed repeats (Alu and LINE-1), and transposable elements is mainly responsible for chromosomal instability and genomic rearrangements. Loss of DNA methylation within heterochromatic regions corresponding to the LOCKs and LADs (exhibiting high DNA methylation levels and association with nuclear membrane in nonneoplastic cells), results in structural reorganization of large heterochromatin blocks and disorganization of the nuclear membrane.