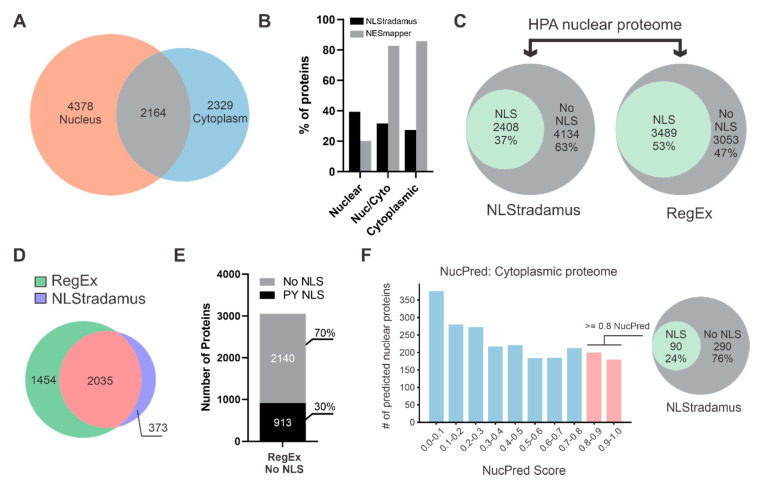
Figure 1.
The majority of human nuclear proteins do not contain a predictable cNLS. (A) Nuclear and cytoplasmic proteins were retrieved from the Human Protein Atlas (HPA) for the identification of distinct nuclear, cytoplasmic and nucleocytoplasmic proteins. (B) cNLS and NES prediction of nuclear, nucleocytoplasmic (Nuc/Cyto) and cytoplasmic proteins from the HPA using NLStradamus and NESmapper, respectively. The majority of Nuc/Cyto and cytoplasmic proteins have a predictable NES in contrast to nuclear and Nuc/Cyto proteins where the majority do not have a predictable cNLS. (C) Comparison of cNLS prediction approaches using NLStradamus and regular expression matching (RegEx) on nuclear proteins from the HPA. All motifs corresponding to cNLSs in the eukaryotic linear motif database were used for RegEx matching, where any protein with at least one match is counted as a hit. Comparing approaches shows that somewhere between 47% and 63% of nuclear proteins do not have a predictable cNLS. (D) The prediction of proteins with a cNLS using either RegEx matching or NLStradamus demonstrates significant overlap. The majority of NLStradamus predictions are also predicted by RegEx matching. (E) Proteins without a cNLS, as determined by RegEx matching, were searched for a minimal PY-NLS (R/H/K-X2-5-PY), demonstrating that a substantial portion of nuclear proteins also do not contain a PY-NLS. (F) Cytoplasmic proteins were analyzed with NucPred to predict nuclear localization and those with a score greater than 0.8 were searched for cNLSs. Those with a cNLS are considered to have a high probability of nuclear localization, highlighting the potential for additional nucleocytoplasmic localizations not supported by the HPA.