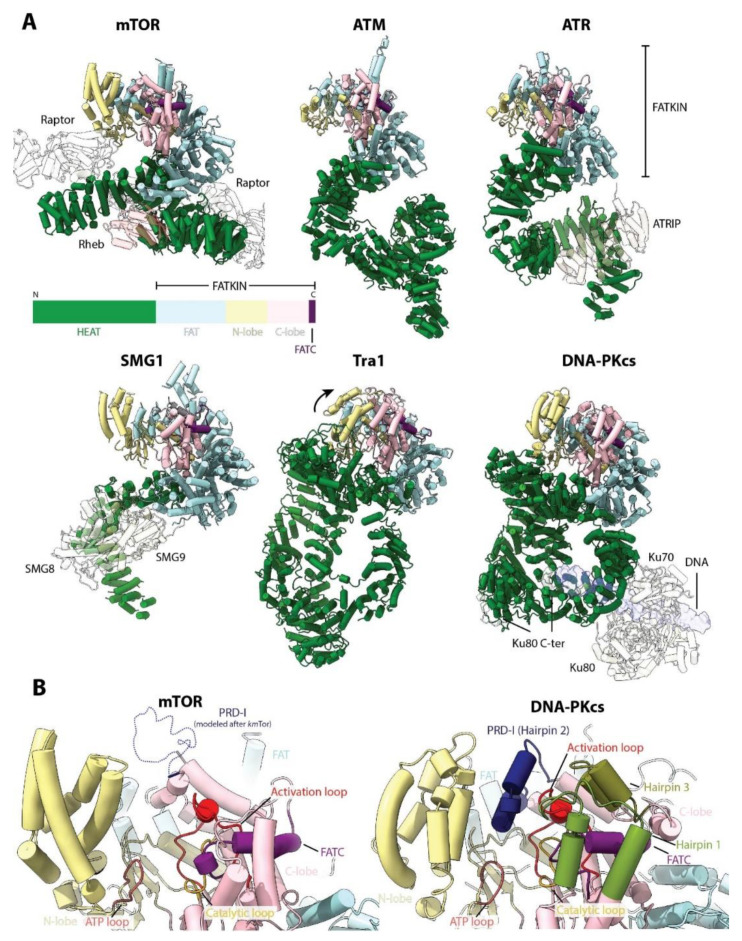
Figure 8.
Compilation of PIKK structures. (A) Comparison of the structures of: H. sapiens mTOR (PDB: 6BCX), H. sapiens ATM (PDB:5NP0), H. sapiens ATR (PDB:5YZ0), H. sapiens SMG1 (PDB:6SYT), S. cerevisiae Tra1 (PDB:5OJS) and H. sapiens DNA-PKcs (PDB:5LUQ and 5Y3R). The conserved domain architecture is indicated schematically (not to scale). All PIKKs share a similar structure, where the N-terminal HEAT domain is the most variable. Other proteins that form part of the complex and/or activate the kinase activity are shown transparent. Tra1 forms part of the bigger SAGA and NuA4 complexes but does not have catalytic activity. For dimeric complexes (mTORC1, ATM), only one monomer is shown for clarity. (B) Comparison of the active site of mTOR (active) and DNA-PKcs (inactive). Conserved elements are indicated. The active site elements can undergo conformational changes in response to upstream cues to further control the activity of the PIKKs.