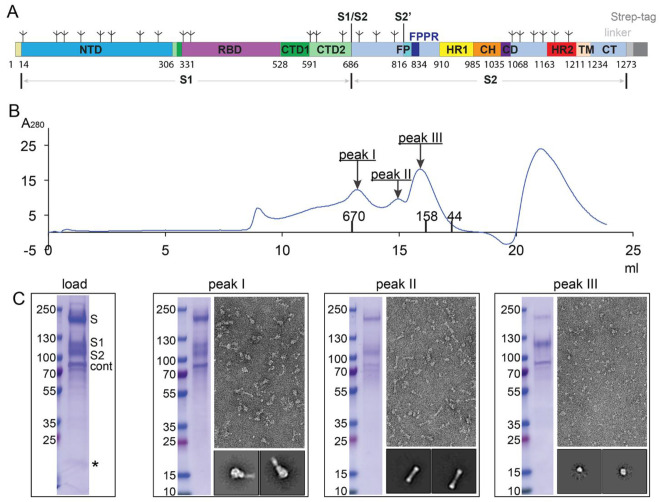
Fig. 1. Preparation of a full-length SARS-CoV-2 spike protein.
(A) Schematic representation of the expression construct of full-length SARS-CoV-2 spike (S) protein. Segments of S1 and S2 include: NTD, N-terminal domain; RBD, receptor-binding domain; CTD1, C-terminal domain 1; CTD2, C-terminal domain 2; S1/S2, S1/S2 cleavage site; S2’, S2’ cleavage site; FP, fusion peptide; FPPR, fusion peptide proximal region; HR1, heptad repeat 1; CH, central helix region; CD, connector domain; HR2, heptad repeat 2; TM, transmembrane anchor; CT, cytoplasmic tail; and tree-like symbols for glycans. A strep-tag was fused to the C terminus of S protein by a flexible linker. (B) The purified S protein was resolved by gel-filtration chromatography on a Superose 6 column in the presence of detergent NP-40. The molecular weight standards include thyoglobulin (670 kDa), γ-globulin (158 kDa) and ovalbumin (44 kDa). Three major peaks (peak I-III) contain the S protein. (C) Load sample and peak fractions from (B) were analyzed by Coomassie stained SDS-PAGE. Labeled bands were confirmed by Western blot (S, S1 and S2) or protein sequencing (S2 and Cont; S and S1 bands did not gave any meaningful results probably due to a blocked N terminus). Cont, copurified contaminating protein, identified as endoplasmic reticulum chaperone BiP precursor by N-terminal sequencing. *, a putative S1/S2-S2’ fragment. Representative images and 2D averages by negative stain EM of three peak fractions are also shown. The box size of 2D averages is ~510Å.