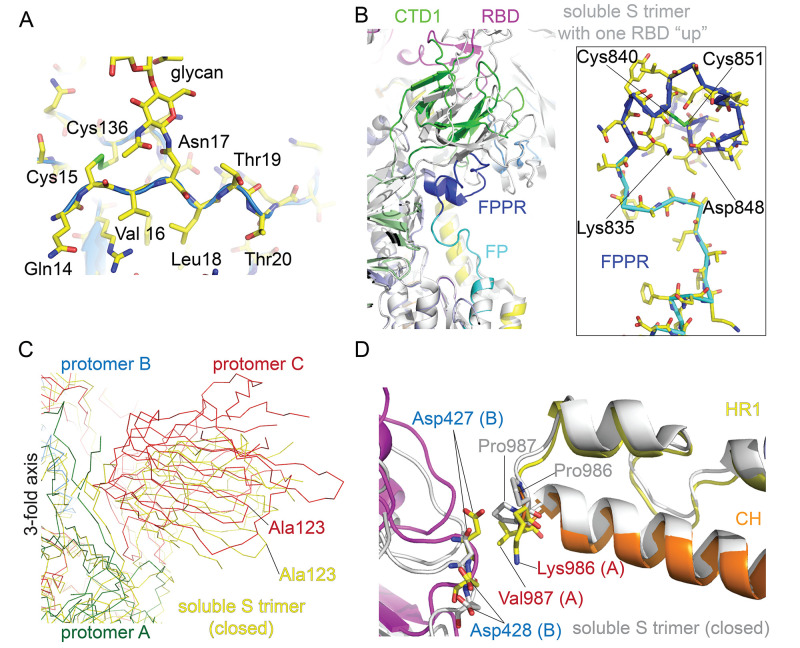
Fig. 3. Selected new features of the SARS-CoV-2 prefusion S trimer.
(A) N-terminal segment of S protein. The N terminus is at residue Gln14 after cleavage of the signal peptide. Cys15 forms a disulfide bond with Cys136. We observed good density for the N-linked glycan at Asn17. (B) A segment immediately downstream of the fusion peptide, while disordered in the stabilized soluble S ectodomain trimer structure, forms a tightly packed structure, designated FPPR for the fusion peptide proximal region, abutting CTD1. The newly identified FPPR structure would clash with CTD1 in the RBD up conformation. Various domains are shown in the color scheme in Fig. 2B. The structure of the soluble S trimer with one RBD in the up conformation (PDB ID: 6vyb) is shown in gray. In the box, a close-up view of the FPPR with adjacent fusion peptide in both surface representation and stick model. (C) The SARS-CoV-2 prefusion S trimer, viewed along the threefold axis, is superposed on the structure of the stabilized soluble S ectodomain trimer in the closed conformation with all three RBDs in the down conformation (PDB ID: 6vxx). While the S2 region is well aligned, there is a significant shift (e.g., ~12Å between two Ala123 residues) in S1. (D) Impact of the proline mutations introduced at residues 986 and 987 to stabilize the prefusion conformation. K986P mutation removes a salt bridge between Lys986 of one protomer and either Asp427 or Asp428 of another protomer in the trimer interface.