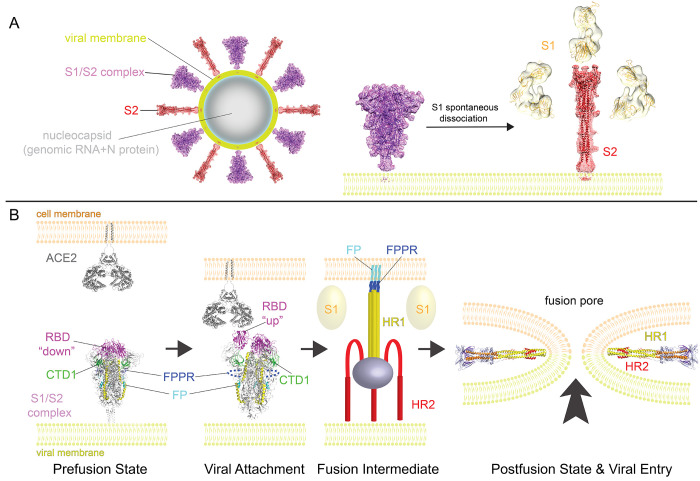
Fig. 5. A model for structural rearrangements of SARS-Cov-2 S protein.
(A) Structural changes independent of a target cell. We suggest that both the prefusion and postfusion spikes are present on the surface of mature virion and the ratio between them may vary (diagram of virion). The postfusion spikes on the virion are formed by S2 after S1 dissociates in the absence of ACE2. (B) ACE2-dependent structural rearrangements. Structural transition from the prefusion to postfusion conformation inducing membrane fusion likely proceeds stepwise as follows: 1) FPPR clamps down RBD through CTD1 in the prefusion S trimer (this study), but it occasionally flips out of position and allows an RBD to sample the up conformation (PDB ID: 6vyb). 2) RBD binding to ACE2 (PBD ID: 6m17) creates a flexible FPPR that enables exposure of the S2’ cleavage site immediately upstream of the adjacent fusion peptide (FP). Cleavage at the S2’ site, and perhaps also the S1/S2 site, releases the structural constraints on the fusion peptide and initiates a cascade of refolding events in S2, probably accompanied by complete dissociation of S1. 3) Formation of the long central three-stranded coiled-coil and folding back of HR2. 4) Formation of the postfusion structure of S2 (this study) that brings the two membranes together, facilitating formation of a fusion pore and viral entry.