Abstract
Soon after the discovery of BRCA1 and BRCA2 over 20 years ago, it became apparent that not all hereditary breast and/or ovarian cancer syndrome families were explained by germline variants in these cancer predisposing genes, suggesting that other such genes have yet to be discovered. BRCA1-associated ring domain (BARD1), a direct interacting partner of BRCA1, was one of the earliest candidates investigated. Sequencing analyses revealed that potentially pathogenic BARD1 variants likely conferred a low–moderate risk to hereditary breast cancer, but this association is inconsistent. Here, we review studies of BARD1 as a cancer predisposing gene and illustrate the challenge of discovering additional cancer risk genes for hereditary breast and/or ovarian cancer. We selected peer reviewed research articles that focused on three themes: (i) sequence analyses of BARD1 to identify potentially pathogenic germline variants in adult hereditary cancer syndromes; (ii) biological assays of BARD1 variants to assess their effect on protein function; and (iii) association studies of BARD1 variants in family-based and case-control study groups to assess cancer risk. In conclusion, BARD1 is likely to be a low–moderate penetrance breast cancer risk gene.
Keywords: BARD1, cancer predisposing gene, hereditary cancer syndromes, breast cancer, ovarian cancer, next-generation sequencing, multi-gene panel testing
1. Introduction
After the discovery of BRCA1 and BRCA2, the highly penetrant breast (BC) and ovarian cancer (OC) predisposing genes, it became evident that about 20% of hereditary BC and/or OC syndrome families did not carry pathogenic variants in these genes. Under the assumption that other genes contribute risk to these cancers, research focused on identifying an additional major risk gene, namely “BRCAX”. Since the discovery of BRCA1 in 1994 [1], 27 cancer predisposing genes that confer significant lifetime risk to hereditary adult cancers have been discovered (reviewed in [2,3]). About half of these genes were discovered using genome-wide linkage analysis to identify candidate loci followed by sequencing candidate genes to identify pathogenic variants segregating in cancer families (reviewed in [2]). However, the rarity of large pedigrees of BRCA1 and BRCA2 mutation negative families and availability of family members suitable for linkage analyses posed considerable challenges for gene discovery projects. With knowledge about gene function, sequencing of candidate genes became a favoured strategy for gene discovery. Candidates can be selected based on having similar biological domains, involvement in the same biological pathway or biochemically interacting with known cancer predisposing gene(s). Using this candidate gene approach over 100 genes have been identified of which the majority have failed to be linked with hereditary cancer syndromes, including BC and/or OC, in independent studies (reviewed in [2,4]).
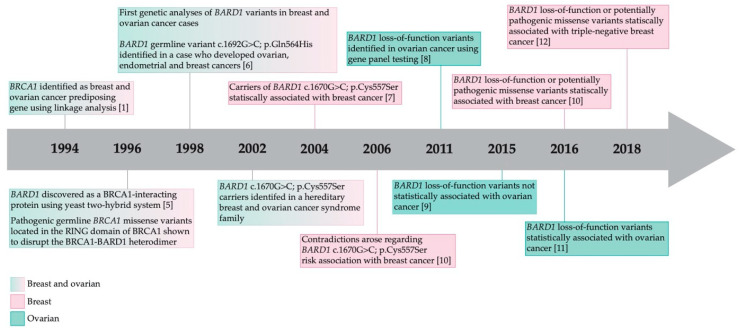
The BRCA1-associated RING domain 1 (BARD1) gene was discovered in 1996 in an effort to understand the biological function of BRCA1 protein as shown in the timeline of major discoveries of BARD1 (Figure 1). BARD1 directly interacts with BRCA1 via its N-terminal RING (really interesting new gene) domain [5]. As pathogenic BRCA1 variants located in the RING domain disrupt BARD1-BRCA1 interaction, it was hypothesized that variants in BARD1 may also affect this interaction, thereby making BARD1 an attractive candidate to pursue in BRCA1 mutation negative cancer families. Although potentially pathogenic BARD1 variants have been reported, the role of BARD1 in cancer predisposition remains inconsistent. Here, we aim to review what is known about BARD1 as a candidate cancer predisposing gene and discuss challenges facing researchers aiming to discover additional cancer predisposing genes. We focused on publications that: (i) described the analyses of potentially pathogenic germline BARD1 variants in adult hereditary cancer syndromes; (ii) BARD1 in vitro and in cellulo assays that assessed the biological effect of potentially pathogenic variants on protein function; and (iii) analyzed cancer risk associated with potentially pathogenic BARD1 variants in family-based and case-control study groups.
Figure 1.
Timeline of the major discoveries associated with BARD1 function and risk to breast and ovarian cancer [1,5,6,7,8,9,10,11,12,13].
2. BARD1 as a Candidate Cancer Predisposing Gene
Little was known about the biological function of BRCA1 when it was first reported as a cancer predisposing gene other than the predicted loss-of-function variants suggested that it may behave as tumour suppressor gene [1]. To learn about BRCA1 function, a yeast two-hybrid screening system was used to identify proteins that directly interact with BRCA1 and a protein was identified that interacts directly with its N-terminal RING domain [5]. Repeating the assay using a human cell line, a similar protein which contained a C-terminal motif with significant homology to the conserved C-terminal BRCA1 C terminus (BRCT) domains in BRCA1 and three tandem ankyrin (ANK) domains adjacent to the BRCT domains was identified. The BRCA1–BARD1 interaction was shown to be disrupted by BRCA1 missense variants [5]. The formation of a stable BRCA1–BARD1 complex was considered to be critical for BRCA1 function and thus BARD1 may play a role as an effector or regulator of BRCA1 [5]. This led to the hypothesis that variants in BARD1 could encode an aberrant protein affecting the interaction with BRCA1 and predispose to BC and/or OC, especially in hereditary cancer syndrome families not accounted for by BRCA1 or BRCA2 [6]. This study was the first report of molecular genetic analyses of BARD1 in cancer samples in 1998 [6] (Figure 1). In a targeted sequencing analysis of 168 BC, OC, and uterine tumours, the first BARD1 variant, c.1692G>C; p.Gln564His, was identified in a woman diagnosed with a clear cell histopathological subtype of OC at the age of 73 years [6]. This variant is predicted to change an amino acid residue located near the BRCT domains in BARD1. This individual was also diagnosed with infiltrating lobular BC and six years later with a clear cell endometrial cancer. Sequencing of cloned BARD1 mRNA transcripts from ovarian and uterine tumour tissues revealed only the variant allele. Sequencing of similarly generated cloned transcripts from non-cancerous uterine tissue revealed both the wild type and variant alleles, suggesting that the missense c.1692G>C; p.Gln564His variant was of germline origin. No family history was available for the variant carrier; thus, it was not possible to determine if this rare variant (not identified in about 300 cancer-free individuals) could be associated with familial cancer. Regardless, the observation that a BARD1 variant was found in an individual with three primary cancers suggests a strong possibility that it is associated with risk to cancer.
Subsequent Studies with Inconsistent Results
The first statistical association was reported by analyzing the BARD1 c.1670G>C; p.Cys557Ser variant in cancer cases from Finnish [7], Nordic [14], and Icelandic [15] populations versus population-matched cancer-free controls. Collectively, these studies were in agreement with the possibility that carriers of this BARD1 variant had an increased risk to BC and/or OC. However, a negative association was reported by three different research groups that analyzed case-control groups from Finnish [8], Australian [16], and Polish [17] populations. The inconsistency in results from the same Finnish population was explained by the possibility of a false positive association due to the small sample sizes [7,8]. Indeed, using publicly available population based genetic databases the minor allele frequency (MAF) of BARD1 c.1670G>C; p.Cys557Ser is 0.015 in the general cancer-free population overall, but it is highly variable among different populations (https://gnomad.broadinstitute.org v2.1.1) [18]. It is the rarest (MAF = 5.2 × 10−5) in East Asians and the most common (MAF = 0.024) in the Ashkenazi Jewish population (Supplementary Table S1). In the Finnish population, this variant has a frequency of approximately 1%, which is relatively more common compared to known pathogenic variants in well-established BC and OC predisposing genes, BRCA1 or BRCA2 (0.001%). This suggests that the effect size of BARD1 c.1670G>C; p.Cys557Ser could be small, that is, it could confer a low–moderate risk to cancer relative to the high risk associated with BRCA1/BRCA2 pathogenic variants.
Some subsequent studies reported no statistically significant association of BARD1 variants with cancer risk, resulting in consideration of BARD1 not being a strong candidate cancer predisposing gene [10,19,20,21]. In part, early studies lacked statistical power due to limited number of carriers identified in the small size of study groups. In 2017, a positive statistical association of BARD1 variants with BC was reported in a large sample size of approximately 30,000 BC cases and 30,000 cancer-free controls [11] (Figure 1). Subsequent studies reported consistent positive results and proposed BARD1 as a candidate cancer predisposing gene where variants confer low–moderate risk for BC, especially for triple-negative BC (TNBC) [13,22,23]. TNBC accounts for 10–20% of all BC cases and is defined by the absence of estrogen and progesterone receptors accompanied by overexpression of human epidermal growth factor receptor 2 (reviewed in [24,25]). These subsequent reports were enriched for TNBC cases, which should have reduced the genetic heterogeneity between BC subtypes [13,22,23,26].
3. Potentially Pathogenic Germline BARD1 Variants Identified in Cancer Cases
3.1. Targeted BARD1 Studies of Breast and Ovarian Cancer
A total of 100 unique germline variants have been reported in studies where targeted analysis of BARD1 was performed (Supplementary Table S1, File S1) [6,7,8,9,10,11,12,14,15,17,24,25,26,27,28,29,30,31,32,33,34]. Variants identified in these BC and OC cases include: 12 frameshift; seven nonsense; five splice site; 67 missense; and nine synonymous variants. These variants are rare in the general population, with the MAFs ranging between 4.2 × 10−7 and 8.12 × 10−1 among cancer-free individuals, though some variants were not identified in this database (https://gnomad.broadinstitute.org v2.1.1) [18]. In BARD1 targeted sequencing studies that investigated ≥500 cancer cases, the allele frequencies ranged between 0.02%–49% [28,33] and 0.03%–48% [10] in BC and OC cases, respectively. Using the American College of Medical Genetics and Genomics (ACMG) guidelines for variant interpretation, only 19 potentially loss-of-function variants are classified as being pathogenic with a large portion of the remaining variants being classified as a variant of uncertain significance. As the pathogenicity of many of these variants is still unclear, we have re-evaluated all of the identified BARD1 missense variants using in silico tools that predict the effect on protein function or conservation at that locus. We have found that 29 of the previously reported missense variants are potentially pathogenic based on being predicted to be damaging by at least five out of seven in silico tools and conserved by all three conservation prediction tools (Supplementary Table S1, File S1). Overall, the mutational spectrum of BARD1 in BC and/or OC is diverse and there does not appear to be any hotspots in any particular domain (Figure 2A,B).
Figure 2.
Potentially pathogenic germline variants reported in the literature mapped to full length BARD1 transcript including RING, ANK and BRCT encoding domains. (A) Predicted loss-of-function variants including nonsense, frameshift and canonical splice site variants. (B) Missense variants predicted to be damaging by in silico tools and conservation tools. (C) Variants discussed in the review. RING = Really Interesting New Gene domain; ANK = Ankyrin domain; BRCT = BRCA1 C Terminus domain. See Supplementary Table S1 for more information.
3.2. Multi-Gene Panel and Next-Generation Sequencing Studies Including BARD1 of Breast and Ovarian Cancers
Multi-gene panels that usually contain known high-risk cancer predisposing genes, such as BRCA1 and BRCA2, were used to determine the prevalence and spectrum of variants in the genes in defined study groups for comparative purposes. Germline whole gene sequencing panels included 10–219 genes [16,18,19,20,21,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62] and whole exome sequencing strategies analyzed 10–832 genes [63,64,65,66,67]. These sequencing strategies were used in studies on BC and/or OC and male BC, the first being published in 2011 [61] (Figure 1). Collectively, 55 unique variants were reported in familial, high-risk or unselected BC and/or OC cases (Figure 2, Supplementary Table S1). Variants identified in these BC and OC cancer cases include: 15 frameshift; 13 nonsense; four splice site; 22 missense; and one synonymous variant. In silico tools revealed that six of the missense variants are predicted to be potentially pathogenic. A germline copy number deletion and deletion of exon 2 were identified in two BC cases [54,65]. A large (1258 bp) heterozygous germline deletion in intron three was identified by multiplex ligation-dependent probe amplification in BARD1 in a BC case (diagnosed at age 36 years) [68]. This deletion was not detected to have an impact on RNA splicing and allelic imbalance could not be measured as there was no SNP to use. These findings are interesting because these variants may result in partial or complete loss of BARD1 protein. In studies of ≥500 cancer cases, the prevalence of potentially pathogenic BARD1 variants ranged from 0.18–0.53% [11,47], 0.67–0.9% [13,22], and 0.07–0.23% [37,63] in BC, TNBC and OC cases, respectively. Clearly, in BC and OC cases carriers of BARD1 variants are much less common than carriers of pathogenic BRCA1/BRCA2 variants (5–10% each).
3.3. BARD1 in Other Cancers
The prevalence of potentially pathogenic germline BARD1 variants has been investigated in colorectal [32,69], endometrial [70] and pancreatic [71,72] cancers (Figure 2A,B, Supplementary Table S1). The carrier frequency of BARD1 in a familial colorectal cancer study was one in 29 (3.4%) [69]. The carrier frequency of BARD1 in unselected endometrial cancer was one in 381 (0.26%) [70] and in two pancreatic cancer studies was one in 302 (0.33%) [71] and nine in 96 (9.4%) [72]. The presence of BARD1 variants in other cancer types suggests that they may also play a role in risk in these diseases. Of the 15 variants that have been identified in these cancer cases, eight have been found in BC and OC cases and two in BC cases.
3.4. Carriers of a BARD1 Variant and a Pathogenic Variant in a Known High-Risk Cancer Predisposing Gene
The carriers of more than one pathogenic variant in different cancer predisposing genes, BRCA1 and BRCA2 as an example, has been described but reports are not common. One in 190,000 Europeans is estimated to carry a pathogenic variant in both BRCA1 and BRCA2 [73,74,75], and this may be higher in defined populations exhibiting founder effects due to common ancestors, such as in the Ashkenazi Jewish (one in 1800) [76,77] and French Canadian [78] populations. Carriers of a BARD1 variant and a pathogenic BRCA1 or BRCA2 variant, that is double heterozygosity, have been reported [14,15,45,48,79].
The contribution of BARD1 variants to cancer risk or disease progression in carriers of pathogenic variants in known cancer predisposing genes is unknown. Four studies of BC cases have investigated the co-occurrence of carriers of the BARD1 c.1670G>C; p.Cys557Ser variant with pathogenic variants in BRCA1 or BRCA2 [14,15,17,80]. The prevalence of this BARD1 variant was significantly higher in Icelandic BC cases who also carried the pathogenic BRCA2 c.771_775del; p.Asn257LysfsTer17 variant known to be a founder pathogenic variant in this population [15]. The risk of BC associated with carrying both variants was estimated to be three-fold higher than carriers of BRCA2 c.771_775del; p.Asn257LysfsTer17 alone and the lifetime probability of developing BC approaches 100% in carriers of both variants. Three other studies of BC cases from Nordic [14], Polish [17] and mixed populations [80] found that carriers of the BARD1 c.1670G>C; p.Cys557Ser variant and a BRCA1 or BRCA2 pathogenic variant did not confirm this finding. Thus, further studies are needed to clarify the interaction of BARD1 with BRCA1 or BRCA2. It is possible that specific BARD1 variants have additive effects by increasing or modifying risk in a linear or synergistic manner in carriers of other high risk variants (reviewed in [81,82,83]).
In our laboratory, whole exome sequencing of 40 BC cases with pathogenic BRCA2 variants identified a carrier of BARD1 c.1339C>G; p.Leu447Val variant (unpublished data). These BC cases were selected by the following criteria: proband with a diagnosis of invasive BC <66 years of age; a strong family history of BC defined by ≥3 BC cases in first-, second- or third-degree relatives in the same familial branch; and all four grandparents originating from Quebec, Canada (French Canadian origin). This rare BARD1 variant, estimated to occur at an overall frequency of 0.006% in the general cancer-free population (https://gnomad.broadinstitute.org) (Supplementary Table S1) [18]. It has been reported twice in the literature and was identified by multi-gene panel testing of 1297 BC cases diagnosed at ≤45 years old [56] and by targeted sequencing of BARD1 in 4469 familial BC cases [33]. Further analysis of other members of our cancer family showed only one of five other carriers was affected by BC (unpublished data). This case is from the French Canadian population and carries a pathogenic variant in BRCA2 c.8537_8538delAG; p.Glu2846GlyfsTer22 known to be a founder pathogenic variant as reported by our group and others [84,85,86,87,88,89,90,91,92,93].
4. The Biological Effect of Potentially Pathogenic BARD1 Variants
4.1. BARD1 Expression in Mammalian Cells
BARD1 is widely expressed in different types of mammalian cells. Early studies in human cell lines demonstrated a constant level of gene expression throughout the cell cycle, in contrast to BRCA1 expression, which has been shown to increase during late G1 phase and reaches a maximum during S phase [94]. Gene expression studies suggest a role of BARD1 in cell growth. Interestingly, partial repression of mouse Bard1 due to antisense RNAs resulted in development of phenotypes characteristic of the early stages of malignancy in murine mammary epithelial cell lines, suggesting a role of Bard1 in tumourigenesis [95]. Moreover, it has been shown that BARD1 encodes multiple protein isoforms in human cells that affect cell growth, and the full length protein is able to suppress tumourigenic phenotypes while alternatively spliced transcripts encode proteins that behave as proto-oncogenes (www.genome.ucsc.edu) (reviewed in [96]). Genetic abnormalities affecting chromosome 2q34–q35 region that harbours the BARD1 locus are not common in BC and OC cases [97,98,99,100]. BARD1 is expressed in breast and ovarian normal tissues as well as in BC and OC tissues (www.proteinatlas.org) and is therefore a plausible candidate cancer predisposing gene.
4.2. BARD1 in Animal and Cell Line Models
Genetically engineered mouse models [101,102] suggest that the mouse orthologue of BARD1 plays a role in tumourigenesis. Human BARD1 and mouse Bard1 orthologues have been shown to share about 70% sequence identity [103]. Bard1−/− knockout mice resulted in embryos that died between days E7.5 and E8.5 due to severe impairment of cell proliferation [101]. Bard1 and p53 double knockout embryos displayed increased chromosomal aneuploidy in comparison to p53−/− mice, suggesting a role in maintaining genomic stability [101]. Bard1+/− heterozygous mice were indistinguishable from their wild-type littermates in terms of viability and fertility and did not develop detectable tumours by 21 months of age. Interestingly, conditional knockouts resulting in the inactivation of Bard1 and/or Brca1 resulted in mice that developed BC with histopathologies strikingly similarity to human TNBC, indicating that this is tumourigenic [102]. The authors suggested that this tumourigenic phenotype could be mediated through the BARD1–BRCA1 heterodimer complex.
4.3. The Biological Impact of Potentially Pathogenic Variants in Different BARD1 Domains
BARD1 protein (777 amino acids; GenBank Accession number CAE48237.1) is structurally similar to BRCA1 (1,863 amino acids; GenBank Accesstion Number AAC37594.1) by having a single N-terminal RING and two C-terminal BRCT domains. However, BARD1 has four tandem ANK domains adjacent to the BRCT domains, which are absent in BRCA1 [5,6,104]. BARD1 maintains genomic stability by functioning in DNA repair, ubiqutination, and transcriptional regulation (reviewed in [105]). The BRCA1–BARD1 complex functions in the homologous recombination DNA repair pathway that repairs DNA double-strand breaks by assisting in the recruitment of RAD51 to the single stranded DNA as a template for repair and downstream pathway function (reviewed in [105]). Aberrant homologous recombination leaves cells prone to genetic alterations and genome instability.
The RING domain is highly conserved and enriched for cysteine residues that are crucial for binding to zinc cations to ensure proper RING domain folding. The RING domain is crucial for direct BRCA1 interaction and stabilization in a cell cycle-dependent manner and subsequently affects DNA repair [5,94,103,106,107,108,109,110]. RING domains are not only found in proteins that exhibit tumour suppressor or proto-oncogenic function but also in proteins that exhibit E3 ubiquitin ligase activity (reviewed in [111,112]). Studies have shown that ubiquitin ligase activity reaches its maximum when BRCA1 is complexed with the RING domain of BARD1 [110,113]. Studies of germline pathogenic BRCA1 variants located in the RING domain disrupted the BRCA1–BARD1 interaction [5]. Other BRCA1 missense variants located in the RING domain impaired E3 ubiquitin ligase activity of the BRCA1–BARD1 heterodimer and affected tumourigenesis [113,114,115]. These findings suggest that having a germline variant in the RING domain of BARD1 could increase risk to BC or OC [6,116]. One frameshift BARD1 variant was identified in a young age of onset BC case (29 years old at diagnosis) by multi-gene panel testing; this variant is predicted to result in premature termination of the protein and partial loss of the RING domain [46]. The BARD1 c.247A>G; p.Cys83Arg protein isoform retained the ability to interact with BRCA1 and retained E3 ubiquitin ligase activity, but the BARD1–BRCA1 complex was less efficient in binding to the nucleosomes and ubiquitylating histone H2A [117]. In contrast, the BARD1 c.253G>T; p.Val85Leu variant had no effect on the BARD1–BRCA1 interaction and homologous recombination DNA repair activity [118]. We classified both variants as potentially pathogenic based on in silico prediction tools (Figure 2B,C, Supplementary Table S1).
Based on amino acid composition BARD1 was thought to contain three ANK repeats but crystallization studies revealed an additional repeat adjacent to the BRCT domain [104]. This newly identified domain may have been missed in earlier studies as it is less conserved than the other ANK domains. These domains are found in a broad spectrum of functionally diverse proteins and have been implicated as sites of highly specific protein-protein interactions (reviewed in [105]). It has been proposed that BARD1 plays a role in apoptosis via its ANK domains in a p53-dependent manner (reviewed in [96,105,119]). Most recently, ANK domains have been shown to be important for recognition of the epigenetic methylation site H4K20me0 on histone H4, for BARD1–BRCA1 recruitment to sites of DNA damage via the homologous recombination DNA repair pathway [120]. Variants in the ANK domains may hinder H4K20me0 recognition which abrogates accumulation of BRCA1 to sites of double-strand breaks, resulting in the activation of alternative DNA repair pathways and genome instability. A deletion of exons 2–6 in BARD1 results in the loss of most of the RING and ANK domains, and consequently its capacity to interact with BRCA1 [121]. Carriers of 14 frameshift and nine nonsense variants located upstream or within the ANK repeats, which could have the effect of encoding a protein lacking ANK domains, have been reported in BC and OC cases (Supplementary Table S1). Interestingly, it was shown that the exonic c.1361C>T; p.Pro454Leu affects the splicing enhancing site, resulting in skipping of exon 5 (Figure 2C) [122], which would result in the loss of a segment of the ANK domains. Studies of BARD1 missense variants, c.1482A>G; p.Asn470Ser and c.1519G>A; p.Val507Met, suggest that these amino acid substitutions did not significantly affect the secondary structure of BARD1 [104]. Indeed, studies have shown that the BARD1 c.1519G>A; p.Val507Met protein isoform has no effect on homologous recombination DNA repair pathway activity [118] (Figure 2C, Supplementary Table S1).
Both C-terminal BRCT domains are well conserved and are often found in proteins that play a role in cell cycle checkpoint and DNA damage response (reviewed in [105]). BRCT domains in BARD1 are recognized by poly (ADP-ribose) polymerase (PARP) as well as other binding proteins [123,124]. Twenty-four frameshift and 15 nonsense variants in BARD1 have been reported upstream or within BRCT domains, which could result in losing these domains fully or partially and could affect function (Figure 2, Supplementary Table S1). It has been shown that the BARD1 c. 1793C>T; p.Thr598Ile protein isoform exhibits significantly lower homologous recombination DNA repair activity [118]. This isoform might affect binding to PARP1, as the amino acid change is predicted to alter the protein’s surface and not any of the known binding sites involved in DNA repair [118].
The biological function of BARD1 variants that lie outside the RING, ANK or BRCT domains have been studied [29,32,125,126,127,128,129,130,131]. The first identified BARD1 c.1692G>C; p.Gln564His variant and the most prevalent c.1670G>C; p.Cys557Ser variant are located in the linker motif between the ANK and BRCT domains (Figure 2C). The BARD1 c.1692G>C; p.Gln564His isoform was shown to disrupt the interaction with the Cleavage stimulating factor 50 (CstF50) protein resulting in impairment of RNA processing. Although both of these isoforms showed no effect on the homologous recombination activity [126], they were found to negatively affect the interaction with p53, resulting in impaired p53-dependent apoptosis [29]. These results suggest that the risk to cancer of these variants is unlikely to be a consequence of defects in homologous recombination DNA repair function but may involve cell cycle or RNA processing pathways.
5. Risk Assessments for Cancer in Carriers of BARD1 Variants
Risk estimates for cancer predisposing genes can be presented as odds ratios (OR) and the risk is interpreted as low (1–1.5), moderate (1.5–5), or high (≥5) [132]. For example, ORs for known BC and OC predisposing genes such as BRCA1 and BRCA2 are high, and variants in these genes have been shown to confer high risk to cancer in carriers. Estimates of the lifetime risk for cancer that are associated with carriers of a potentially pathogenic BARD1 variant have been reported for BC and OC in case-control studies, although most involved familial BC cases. Risk estimates have been determined for individual variants or all variants found in the gene (gene-based risk) (Table 1 and Table 2).
Table 1.
Risk of breast or ovarian cancer associated with BARD1.
| Reference | Cancer Type | Selection of Cancer Cases | Number of Cases | Number of Controls | Population | Type of Variants Included | OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|---|---|
| [11] | Breast | Unselected | 28,536 | 26,078 b | Mixed e | LoF or missense f | 2.16 (1.31–3.63) | 2.26 × 10−3 |
| [52] | Breast | Familial | 2134 | 26,375 b | Mixed e | LoF or missense g | 3.18 (1.34–7.36) | 1.22 × 10−2 |
| [13] | TNBC | High risk | 4090 | 26,079 b | White | LoF or missense f | 5.92 (3.36–10.27) | 2.20 × 10−9 |
| TNBC | High risk | 2003 | 26,079 b | White | LoF or missense f | 4.35 (2.02–9.30) | 7.6 × 10−4 | |
| [23] | Breast | Familial | 3667 | – b | French | LoF or missense g | 2.00 (0.74–4.10) | – |
| TNBC | Familial | 3667 | – b | French | LoF or missense g | 11.27 (3.37–25.01) | – | |
| [33] | Breast | Familial | 4469 | 37,265 c | Unknown | LoF or missense f | 5.35 (3.17–9.04) | <10−5 |
| [22] | TNBC | Unselected | 4824 | 123,136 d | Mixed e | LoF or missense f | 9.76 (6.77–13.87) | – |
| [60] | Ovarian a | Unselected | 1915 | 36,276 b | Mixed e | LoF or missense g | 4.2 (1.4–12.5) | 0.02 |
| [63] | Ovarian a | High risk | 4122 | 4688 | White | LoF | 1.59 (0.31–9.56) h | 0.57 |
TNBC = triple-negative breast cancer; LoF = loss-of-function variant (nonsense, frameshift or canonical splice site); OR = odds ratio; 95% CI = 95% confidence interval; ExAC = Exome Aggregation Consortium; FLOSSIES = Fabulous Ladies Over Seventy; gnomAD = Genome Aggregation Database; ACMG = American College of Medical Genetics and Genomics [136,137]; – = not stated; a Mixed histopathological subtypes; b ExAC; c ExAC, FLOSSIES, and geographically matched female controls; d gnomAD; e Majority of cases are white; f ACMG; g Missense variants were classified using different bioinformatic tools in each study; h Adjusted OR was calculated as 6.3 (95%CI 0.55–74.25, p = 0.19).
Table 2.
Risk of breast or ovarian cancer associated with potentially pathogenic BARD1 variants.
| Reference | Coding DNA Reference Sequence a | Predicted Amino Acid Change | Cancer Type | Selection of Cancer Cases | Number of Cases | Number of Controls | Population | OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|---|---|---|
| [28] | c.70C>T | p.Pro24Ser | Breast | Unselected | 507 | 539 | China | 0.68 (0.52–0.9) | – |
| [34] | c.722C>G | p.Ser241Cys | Breast | Unselected | 143 | 155 | Japan | 1.57 (0.61–4.05) b | – |
| [34] | c.1139_1159del | p.Asn380_Phe387delinsIle | Breast | Unselected | 143 | 155 | Japan | 0.96 (0.49–1.86) c | – |
| [34] | c.1134G>C | p.Arg378Ser | Breast | Unselected | 143 | 155 | Japan | 1.35 (0.7–2.61) d | – |
| [28] | c.1134G>C | p.Arg378Ser | Breast | Unselected | 507 | 539 | China | 0.94 (0.72–1.24) | – |
| [26] | c.1972C>T | p.Arg658Cys | Breast | Unselected | 12,476 | 4707 | Poland | 1.16 (0.75–1.81) | 0.51 |
| [26] | c.1972C>T | p.Arg658Cys | TNBC | Unselected | 1120 | 4707 | Poland | 0.97 (0.40–2.36) | 0.95 |
| [26] | c.1977A>G | p.Arg659= | Breast | Unselected | 12,476 | 4707 | Poland | 1.32 (0.73–2.40) | 0.36 |
| [26] | c.1977A>G | p.Arg659= | TNBC | Unselected | 1120 | 4707 | Poland | 0.60 (0.14–2.64) | 0.5 |
| [34] | c.1518C>T | p.His506= | Breast | Unselected | 143 | 155 | Japan | 0.71 (0.36–1.42) e | – |
| [34] | c.1519G>A | p.Val507Met | Breast | Unselected | 143 | 155 | Japan | 1.28 (0.8–2.04) f | – |
| [8] | c.1519G>A | p.Val507Met | Breast | Familial | 663 | 718 | Finland | 1.04 (0.8–1.34) | 0.79 |
| [8] | c.1519G>A | p.Val507Met | Breast | Unselected | 867 | 718 | Finland | 1.27 (0.99–1.63) | 0.06 |
| [28] | c.1519G>A | p.Val507Met | Breast | Unselected | 507 | 539 | China | 0.98 (0.75–1.29) | – |
| [7] | c.1670G>C | p.Cys557Ser | Breast or ovarian | Familial | 126 | 1018 | Finland | 4.2 (1.7–10.7) | 5 × 10−3 |
| [8] | c.1670G>C | p.Cys557Ser | Breast | Familial | 926 | 725 | Finland | 0.49 (0.24–1.01) | 0.05 |
| [8] | c.1670G>C | p.Cys557Ser | Breast | Familial | 255 | 358 | Finland | 0.70 (0.21–2.34) | 0.56 |
| [8] | c.1670G>C | p.Cys557Ser | Breast | Unselected | 868 | 725 | Finland | 0.74 (0.38–1.44) | 0.38 |
| [8] | c.1670G>C | p.Cys557Ser | Breast | Unselected | 697 | 358 | Finland | 1.09 (0.47–2.56) | 0.84 |
| [14] | c.1670G>C | p.Cys557Ser | Breast or ovarian | Familial | 757 | 3591 | Nordic | 1.9 (1.32–2.83) | 10−3 |
| [14] | c.1670G>C | p.Cys557Ser | Breast | Unselected | 1984 | 3591 | Nordic | 1.7 (1.23–2.22) | 10−3 |
| [15] | c.1670G>C | p.Cys557Ser | Breast | Unselected | 992 | 703 | Iceland | 1.82 (1.11–3.01) | 0.014 |
| [17] | c.1670G>C | p.Cys557Ser | Breast | High risk | 3188 | 1038 | Poland | 1.2 (0.9–1.7) | 0.3 |
| [16] | c.1670G>C | p.Cys557Ser | Breast | Unselected | 1136 | 623 | Australia | 0.80 (0.50–1.27) g | 0.3 |
| [35] | c.1670G>C | p.Cys557Ser | Breast | High risk | 322 | 570 | Chile | 1.4 (0.6–3.7) | 0.47 |
| [64] | c.1690C>T | p.Gln564Ter | Breast | Familial | 1018 | 2036 | Poland | 1.0 (0.1–8.6) | 1 |
| [26] | c.1690C>T | p.Gln564Ter | Breast | Unselected | 13,935 | 5896 | Poland, Belarus | 2.30 (1.03–5.15) h | 0.04 |
| [26] | c.1690C>T | p.Gln564Ter | TNBC | Unselected | 1120 | 4707 | Poland | 3.62 (1.21–10.78) | 0.02 |
TNBC = triple-negative breast cancer; OR = odds ratio; 95% CI = 95% confidence interval; – = not stated; a Human GRCh37/hg19 transcript NM_000465.4; OR adjusted for age, first-degree family history of breast cancer, age at first live birth and body mass index was calculated as b 1.65 (95%CI 0.61–4.43), c 0.94 (95%CI 0.46–1.90), d 1.38 (95%CI 0.69–2.76), e 0.67 (95%CI 0.32–1.41), f 1.32 (95%CI 0.81–2.16); g Adjusted OR is shown as the crude OR was not reported; h OR adjusted for study origin was 2.24 (95%CI 0.99–5.03, p = 0.05) and using the Mantel-Haenszel method was 2.12 (95%CI 0.97–4.62, p = 0.06).
In our literature review, we found three studies that had reported risk estimates for BC, where they were derived from BC cases not selected for family history of cancer [11,22,60] (Table 1). In these studies the ORs for the association of potentially pathogenic BARD1 variants and risk for BC (any subtype), TNBC or OC were estimated as 2.16 (95% confidence interval (CI) 1.31–3.63, p = 2.26 × 10−3), 9.76 (95% CI 6.77–13.87, p <0.05) and 4.2 (95% CI 1.4–12.5, p = 0.02), respectively. The narrow CI for any subtype of BC reported by Couch et al. [11], is a reflection of the large sample size of BC cases (N = 28,536) compared with the other studies. In one study the estimated risk was higher for BC to be diagnosed before 40 years of age in familial cases (OR 12.04, 95% CI 5.78–25.08, p <0.001) [33], which is consistent with younger age of diagnosis being associated with carriers of pathogenic variants in known cancer predisposing genes, such as BRCA1 [133]. A meta-analysis of risk estimates for 37 known or proposed genetic risk factors estimated that the OR for BARD1 was 2.33 for BC and <2 for OC [134].
A number of studies estimated the cancer risk associated with specific variants in BARD1 in BC and hereditary BC and/or OC syndrome families (Table 2). We found reports describing the risk for nine different BARD1 variants: one nonsense, one frameshift, six missense and two synonymous variants. Risk estimates are variable across these studies, where the highest risk was observed for c.1690C>T; p.Gln564Ter in association with TNBC cases not selected for family history of cancer (OR 3.62, 95% CI 1.21–10.78, p = 0.02). This is consistent with the previous report suggesting that BARD1 variants may be associated with a higher risk for TNBC [22]. A meta-analysis of the c.1670G>C; p.Cys557Ser variant in 11,870 BC cases and 7687 controls, from Nordic, Australian, Canadian, Russian, English, and Polish populations, concluded that carriers of this BARD1 variant have a low risk for BC (OR 1.14, 95%CI 0.94–1.34, p = 0.13) [135]. While studies estimating the familial risk of BARD1 variants to OC are still lacking, evidence is emerging that BARD1 is a potential moderate risk gene for TNBC.
6. BARD1 in Medical Genetics Settings
Although risk estimates for developing cancer in carriers of BARD1 variants remains to be validated for clinical settings, BARD1 has been included in multi-gene testing panels in hereditary cancer medical genetic settings. Indeed, BARD1 was added to gene testing panels not long after it was proposed as a cancer predisposing gene. Given that risk for cancer in carriers of BARD1 variants is equivocal, we questioned if there are recommendations should potentially pathogenic variants be identified by panel testing. We reviewed publicly available resources for statements akin to those providing guidance for the genetic counselling of carriers of known high risk cancer predisposing genes (Table 3). Recommendations for the known high risk genes, BRCA1 and BRCA2, include increased breast awareness and screening, discussion of risk-reducing mastectomy and/or bilateral-salpingo oophorectomy, education about cancer signs and symptoms and genetic counselling (see The National Comprehensive Cancer Network (NCCN) Guidelines for Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic Cancer, Version 1.2020). The NCCN guidelines for moderate risk BC genes state that there is an increased risk of female BC with management options for increased screening (mammography), but not risk-reducing mastectomy as there is insufficient evidence. American Society of Clinical Oncology (ASCO) guidelines [138] and the ACMG statement [139] for BC recommend that for moderate-penetrance genes the mutation status should not directly alter treatment, mastectomy or systemic therapy options as data is lacking for these genes. The NCCN Guidelines added BARD1 to their 2020 version stating that though there is a potential for increased risk for BC in female heterozygous carriers of BARD1 variants (risk not defined), though there is insufficient evidence to make recommendations (www.nccn.org). The risk for other cancers, including OC, is insufficient to impact risk management. The other resources (ASCO, ACMG, and Institut national d’excellence en santé et en services sociaux (INESSS), a Quebec, Canada resource) did not mention BARD1. Nonetheless, BARD1 continues to be included in multi-gene testing panels for clinical purposes, including commercially available resources. It has been argued that the continued testing of BARD1 would allow for ready access to carrier status if and when risk estimates and recommendations become available in the future.
Table 3.
Publicly available resources reviewed for recommendations for management of carriers of a potentially pathogenic BARD1 variant.
| Reference | Resource Name | Acronym | Resource Type | BARD1 Included in Resource |
|---|---|---|---|---|
| www.nccn.org | National Comprehensive Cancer Network | NCCN | Guideline | Yes |
| [138,140] | American Society of Clinical Oncology | ASCO | Guideline | No |
| [139] | American College of Medical Genetics and Genomics | ACMG | Statement | No |
| www.inesss.qc.ca | Institut national d’excellence en santé et en services sociaux | INESSS | Statement | No |
Clinical diagnostic laboratories as well as researchers have used the ClinVar database [141] (www.ncbi.nlm.nih.gov/clinvar) to access reports of variants identified in human diseases, including those for BARD1. Submissions to ClinVar are accepted from a number of different groups but the majority come from clinical testing laboratories and very few variants have been assessed for their biological effect on protein function. Each variant is interpreted and assigned a clinical significance: benign, likely benign, uncertain significance, likely pathogenic or pathogenic. There have been a number of reports of BARD1 variants in the context of hereditary cancer syndromes: 160 pathogenic, 19 pathogenic or likely pathogenic, 56 likely pathogenic, and 1131 variants of uncertain significance (reviewed in [96]). We suggest that ClinVar should be used with caution as a resource to determine if a BARD1 variant may play a role in hereditary cancer risk, as few of these variants have been vetted by expert review panels akin to those available for variants in BRCA1 and BRCA2 (www.brcaexchange.org) [142].
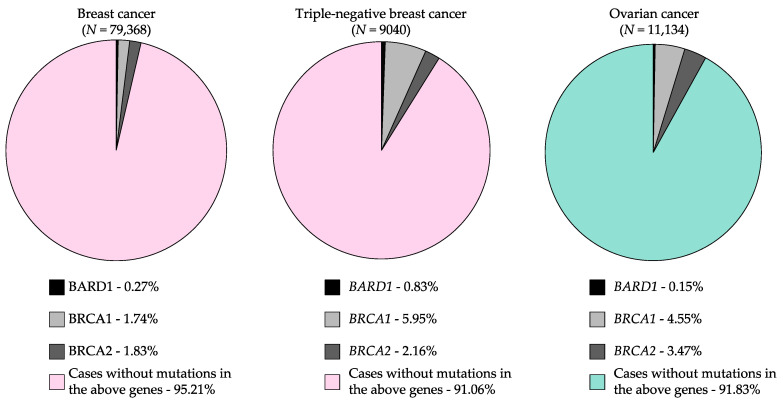
The prevalence of variants in high risk cancer predisposing genes in clinical genetic testing settings has been difficult to estimate due to lack of access to data. Recently, Ambry Genetics® (which offers commercially available and clinically approved multi-gene panel tests) developed an open access web-based tool to query prevalence of pathogenic variants in 49 genes, including BARD1, found in nine cancer types (www.ambrygen.com/clinician/resources/prevalence-tool) [143]. Pathogenic variants in BARD1 were identified in 0.27% of BC cases and 0.15% of OC cases (Figure 3, Table 4). Although there is a higher prevalence of carriers in BC cases with a family history of BC and/or OC than those with no family history of cancer, findings are not statistically significant. The prevalence of carriers was also higher in OC cases with a family history of BC than those not selected for family history of cancer, though this difference is not statistically significant. The highest prevalence of carriers was found in TNBC cases (0.85%)—a subtype of BC aforementioned to be most likely to be associated with moderate risk in carriers of BARD1 variants—which is significantly higher than BC cases unselected for subtype (p < 0.001) (Figure 3, Table 4).
Figure 3.
Proportion of carriers of pathogenic BARD1 variants in breast cancer and ovarian cancer cases relative to carriers of variants in known cancer predisposing genes, BRCA1 and BRCA2. Source: Interactive Prevalence Tables From Multi-Gene Panel Testing: A collaboration between investigators from Mayo Clinic and Ambry Genetics® [143] (www.ambrygen.com/clinician/resources/prevalence-tool).
Table 4.
Prevalence of BARD1 variants in breast and ovarian cancer cases a.
| Context | Number of Cases Tested | Number of Carriers | % |
|---|---|---|---|
| Personal history of breast cancer | 59,375 | 158 | 0.27 |
| ER+ | 30,950 | 51 | 0.16 |
| PR+ | NA | NA | NA |
| HER2+ | 1670 | 4 | 0.24 |
| TNBC | 6745 | 56 | 0.83 |
| Family history of cancer | |||
| Breast | 37,372 | 107 | 0.29 |
| Ovarian | 7990 | 20 | 0.25 |
| Breast and ovarian | 5138 | 18 | 0.35 |
| No cancer | 3491 | 8 | 0.23 |
| Personal history of ovarian cancer | 9149 | 14 | 0.15 |
| Family history of cancer | |||
| Breast | 3867 | 9 | 0.23 |
| Ovarian | NA | NA | NA |
| Breast and ovarian | NA | NA | NA |
| No cancer | NA | NA | NA |
ER = estrogen receptor; PR = progesterone receptor; HER2 = human epidermal growth factor receptor 2; TNBC = triple-negative breast cancer. a Source: Interactive Prevalence Tables From Multi-Gene Panel Testing: A collaboration between investigators from Mayo Clinic and Ambry Genetics® [143] (www.ambrygen.com/clinician/resources/prevalence-tool).
7. Our Perspective of BARD1 as a Cancer Predisposing Gene
Since the discovery over 20 years ago that BARD1 interacts with BRCA1, numerous studies have investigated the possibility that BARD1 variants may affect risk to the heritable form of BC and OC, and perhaps account for the missing heritability not attributed to BRCA1/BRCA2, historically denoted as “BRCAX”. However, genetic studies involving family-based cancer cases and case-control study groups have consistently shown that potentially pathogenic variants in BARD1 are unlikely to: i) confer a high lifetime risk (OR ≥ 5) to BC and OC in heterozygous carriers akin to pathogenic variants described for BRCA1 and BRCA2; and ii) account for the remaining or a significant proportion of heritability of BC and OC attributable to known cancer predisposing genes, especially in cancer syndromes involving these cancer types. Indeed, such studies investigating diverse populations worldwide support the hypothesis that potentially pathogenic variants in BARD1 confer a low–moderate risk to cancer, with perhaps a moderate risk to TNBC. Molecular biology studies of BARD1 support a role in neoplasia in modifying cell growth and tumour suppressor activity leading to tumourigenesis, particularly in the context of modelling variants found associated with heritable disease. Although collectively the research literature supports a role for BARD1 variants in the etiology of cancer, there is insufficient evidence to translate findings to clinical settings for the purposes of providing cancer risk in variant carriers or managing cancer care as is currently available for carriers of variants in known high risk genes, such as BRCA1 and BRCA2 [144].
There are barriers to estimating risk for BARD1 variants: i) the rarity of carriers of potentially pathogenic BARD1 variants; ii) the posited low–moderate risk of cancer associated with BARD1 variants; and iii) the heterogeneity within and between populations, cancer cases of the same primary tissue site and cancer subtypes (based on histopathology or biomarkers). A family-based study estimated that the relative risk associated with BARD1 carriers was 2.27 (95%CI 0.47–18.91) for BC [47], suggesting that women with a family history of BC have at least a two-fold increased risk for this disease as compared with those who do not. However, the wide confidence interval, reflecting small sample groups in this study, suggest that this information should be viewed with caution. It will be difficult to identify a sufficiently large sample size to improve risk estimates as the numbers of carriers of BARD1 variants are rare, as has been observed for carriers of potentially pathogenic PALB2, BRIP1, RAD51C or RAD51D variants in hereditary BC and/or OC cancer syndrome families (Figure 3) [145,146]. For PALB2 the risk of a pathogenic variant was estimated with high confidence in an international study of 524 families [146]. It has been estimated that approximately 40,000 OC cases and 40,000 controls would be required to estimate risk for BARD1 variant carriers, should this risk be in the low-moderate range [10]. Ideally, the appropriate control population should be matched with the cases on age and sex and have no family history of cancer. Cancer cases should have clearly defined histopathology to more precisely characterize associations with TNBC (Table 1). The design of future studies to assess the risk of BARD1 pathogenic variants in different cancers will be crucial to determine if translation to patient care in the clinic is appropriate.
A number of BARD1 missense variants have been characterized and shown to directly alter BARD1 function or its interaction with BRCA1 (Figure 2). It will be important to continue to functionally characterize variants, especially missense variants identified through hereditary cancer studies and clinics, to assess risk. Evaluating new nonsense variants in the C-terminus of the protein will also be important, as previous studies have shown with BRCA2 c.9976A>T; p.Lys3326Ter that truncation of the end of protein may not significantly increase risk to BC [141,147]. Moreover, in cellulo functional assays have demonstrated that the impact of some nonsense variants are not necessarily associated with nonsense-mediated decay [148]. Canonical splicing variants will also be important to assess as aberrant splicing is known to affect the expression of BARD1 and may lead to mis-localization of the protein (reviewed in [96]). Furthermore, studies are lacking that specifically address genotype-phenotype correlations, variable expressivity, epigenetic modification(s), somatic mosaicism, large genomic rearrangements or de novo germline variants. As BARD1 variants have also been identified in carriers of known cancer predisposing genes, BRCA1 and BRCA2, it will be important to explore if BARD1 variants modify risk in an epistatic or additive manner in this context.
Genetic epidemiology studies have identified a number of host factors that modify cancer risk regardless of variant carrier status (reviewed in [149]). For example, it has been shown that carriers of pathogenic BRCA1 or BRCA2 variants have reduced risk for OC due to oral contraceptive pill use or parity or surgical intervention (i.e., tubal ligation or oophorectomy) [150,151,152,153,154]. As the risk attributed to potentially pathogenic BARD1 variants is expected to be low–moderate, interactions with these host factors should be investigated; results could inform the development of guidelines for patient management and genetic counselling.
The clinical impact of BARD1 dysfunction in cancer cells is currently unknown. Carriers of pathogenic BRCA1 or BRCA2 variants respond well to platinum chemotherapies [155,156] and to new therapies such as PARP inhibitors that take advantage of DNA repair pathways affected by pathogenic variants in these genes [157,158,159,160,161,162]. Assaying BARD1 variants in women with BC and OC as biomarkers for guiding therapeutic use has not been fully explored.
Evidence from the literature presented in this review is sufficient to conclude that variants in BARD1 can confer low–moderate risk to BC, especially TNBC, or modify risk to BC in carriers of pathogenic BRCA1 or BRCA2 variants (so-called double heterozygous carriers). However, much work remains, and continued research with larger well-defined study subjects with associated clinical correlates, with additionally discovered variants that have been characterized in biological assays, will elucidate the role of BARD1 in the etiology of cancer and help develop guidelines for care in carriers and new therapies.
Acknowledgments
The authors acknowledge Corinne Serruya and Mary Fujiwara for assistance in editing drafts and conceptualization. The authors acknowledge the National Comprehensive Cancer Network guidelines for information regarding clinical management of carriers of variants in cancer predisposing genes. The authors acknowledge Evan Weber, Zaki El Haffaf for discussions about clinical guidelines and recommendations, and Karine Bédard for assistance with statements.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/8/856/s1, Supplementary Table S1: All BARD1 genetic variants identified from a literature review; Supplementary file S1: Methodology.
Author Contributions
Conceptualization, W.M.A., C.T.F., and P.N.T.; writing—original draft preparation, W.M.A., C.T.F. and N.R.; writing—review and editing, P.N.T.; supervision, P.N.T.; All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by: The Canadian Institute for Health Research (CIHR) operating grants [PCC-156736 to P.N.T., Celia M.T. Greenwood, Jiannis Ragoussis; PJT-156124 to P.N.T., Jiannis Raggousis]; The RI-MUHC and CRCHUM receive support from the FRQS. W.M.A. is supported by a Taibah University Scholarship Award and The Ministry of Higher Education, Saudi Arabia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P.A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L.M., Ding W., et al. Strong Candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Rahman N. Realizing the promise of cancer predisposition genes. Nature. 2014;505:302–308. doi: 10.1038/nature12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fearon E.R. Human cancer syndromes: Clues to the origin and nature of cancer. Science. 1997;278:1043–1050. doi: 10.1126/science.278.5340.1043. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen F.C., van Overeem Hansen T., Sørensen C.S. Hereditary breast and ovarian cancer: New genes in confined pathways. Nat. Rev. Cancer. 2016;16:599–612. doi: 10.1038/nrc.2016.72. [DOI] [PubMed] [Google Scholar]
- 5.Wu L.C., Wang Z.W., Tsan J.T., Spillman M.A., Phung A., Xu X.L., Yang M.C., Hwang L.Y., Bowcock A.M., Baer R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat. Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 6.Thai T.H., Du F., Tsan J.T., Jin Y., Phung A., Spillman M.A., Massa H.F., Muller C.Y., Ashfaq R., Mathis J.M., et al. Mutations in the BRCA1-associated RING domain (BARD1) gene in primary breast, ovarian and uterine cancers. Hum. Mol. Genet. 1998;7:195–202. doi: 10.1093/hmg/7.2.195. [DOI] [PubMed] [Google Scholar]
- 7.Karppinen S.-M., Heikkinen K., Rapakko K., Winqvist R. Mutation screening of the BARD1 gene: Evidence for involvement of the Cys557Ser allele in hereditary susceptibility to breast cancer. J. Med. Genet. 2004;41:e114. doi: 10.1136/jmg.2004.020669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karppinen S.-M., Barkardottir R.B., Backenhorn K., Sydenham T., Syrjäkoski K., Schleutker J., Ikonen T., Pylkäs K., Rapakko K., Erkko H., et al. Nordic collaborative study of the BARD1 Cys557Ser allele in 3956 patients with cancer: Enrichment in familial BRCA1/BRCA2 mutation-negative breast cancer but not in other malignancies. J. Med. Genet. 2006;43:856–862. doi: 10.1136/jmg.2006.041731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stacey S.N., Sulem P., Johannsson O.T., Helgason A., Gudmundsson J., Kostic J.P., Kristjansson K., Jonsdottir T., Sigurdsson H., Hrafnkelsson J., et al. The BARD1 Cys557Ser variant and breast cancer risk in Iceland. PLoS Med. 2006;3:e217. doi: 10.1371/journal.pmed.0030217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vahteristo P., Syrjäkoski K., Heikkinen T., Eerola H., Aittomäki K., von Smitten K., Holli K., Blomqvist C., Kallioniemi O.-P., Nevanlinna H. BARD1 variants Cys557Ser and Val507Met in breast cancer predisposition. Eur. J. Hum. Genet. 2006;14:167–172. doi: 10.1038/sj.ejhg.5201542. [DOI] [PubMed] [Google Scholar]
- 11.Johnatty S.E., Beesley J., Chen X., Hopper J.L., Southey M.C., Giles G.G., Goldgar D.E., Chenevix-Trench G., Spurdle A.B. The BARD1 Cys557Ser polymorphism and breast cancer risk: An Australian case-control and family analysis. Breast Cancer Res. Treat. 2009;115:145–150. doi: 10.1007/s10549-008-0045-y. [DOI] [PubMed] [Google Scholar]
- 12.Jakubowska A., Cybulski C., Szymańska A., Huzarski T., Byrski T., Gronwald J., Debniak T., Górski B., Kowalska E., Narod S.A., et al. BARD1 and breast cancer in Poland. Breast Cancer Res. Treat. 2008;107:119–122. doi: 10.1007/s10549-007-9537-4. [DOI] [PubMed] [Google Scholar]
- 13.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141, 456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guénard F., Labrie Y., Ouellette G., Beauparlant C.J., Durocher F. Genetic sequence variations of BRCA1-interacting genes AURKA, BAP1, BARD1 and DHX9 in French Canadian families with high risk of breast cancer. J. Hum. Genet. 2009;54:152–161. doi: 10.1038/jhg.2009.6. [DOI] [PubMed] [Google Scholar]
- 15.Ramus S.J., Song H., Dicks E., Tyrer J.P., Rosenthal A.N., Intermaggio M.P., Fraser L., Gentry-Maharaj A., Hayward J., Philpott S., et al. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J. Natl. Cancer Inst. 2015;107 doi: 10.1093/jnci/djv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Couch F.J., Hart S.N., Sharma P., Toland A.E., Wang X., Miron P., Olson J.E., Godwin A.K., Pankratz V.S., Olswold C., et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J. Clin. Oncol. 2015;33:304–311. doi: 10.1200/JCO.2014.57.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorringe K.L., Choong D.Y.H., Visvader J.E., Lindeman G.J., Campbell I.G. BARD1 variants are not associated with breast cancer risk in Australian familial breast cancer. Breast Cancer Res. Treat. 2008;111:505–509. doi: 10.1007/s10549-007-9799-x. [DOI] [PubMed] [Google Scholar]
- 18.Couch F.J., Shimelis H., Hu C., Hart S.N., Polley E.C., Na J., Hallberg E., Moore R., Thomas A., Lilyquist J., et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 2017;3:1190–1196. doi: 10.1001/jamaoncol.2017.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu C., Polley E.C., Yadav S., Lilyquist J., Shimelis H., Na J., Hart S.N., Goldgar D.E., Shah S., Pesaran T., et al. The contribution of germline predisposition gene mutations to clinical subtypes of invasive breast cancer from a clinical genetic testing cohort. J. Natl. Cancer Inst. 2020 doi: 10.1093/jnci/djaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimelis H., LaDuca H., Hu C., Hart S.N., Na J., Thomas A., Akinhanmi M., Moore R.M., Brauch H., Cox A., et al. Triple-Negative breast cancer risk genes identified by multigene hereditary cancer panel testing. J. Natl. Cancer Inst. 2018;110:855–862. doi: 10.1093/jnci/djy106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castéra L., Harter V., Muller E., Krieger S., Goardon N., Ricou A., Rousselin A., Paimparay G., Legros A., Bruet O., et al. Landscape of pathogenic variations in a panel of 34 genes and cancer risk estimation from 5131 HBOC families. Genet. Med. 2018;20:1677–1686. doi: 10.1038/s41436-018-0005-9. [DOI] [PubMed] [Google Scholar]
- 22.Kumar P., Aggarwal R. An overview of triple-negative breast cancer. Arch. Gynecol. Obstet. 2016;293:247–269. doi: 10.1007/s00404-015-3859-y. [DOI] [PubMed] [Google Scholar]
- 23.Ryu D.W., Jung M.J., Choi W.S., Lee C.H. Clinical significance of morphologic characteristics in triple negative breast cancer. J. Korean Surg. Soc. 2011;80:301. doi: 10.4174/jkss.2011.80.5.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suszynska M., Kluzniak W., Wokolorczyk D., Jakubowska A., Huzarski T., Gronwald J., Debniak T., Szwiec M., Ratajska M., Klonowska K., et al. BARD1 is a low/moderate breast cancer risk gene: Evidence Based on an association study of the central European p.Q564X Recurrent mutation. Cancers (Basel) 2019;11:740. doi: 10.3390/cancers11060740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratajska M., Antoszewska E., Piskorz A., Brozek I., Borg Å., Kusmierek H., Biernat W., Limon J. Cancer predisposing BARD1 mutations in breast-ovarian cancer families. Breast Cancer Res. Treat. 2012;131:89–97. doi: 10.1007/s10549-011-1403-8. [DOI] [PubMed] [Google Scholar]
- 26.Huo X., Hu Z., Zhai X., Wang Y., Wang S., Wang X., Qin J., Chen W., Jin G., Liu J., et al. Common non-synonymous polymorphisms in the BRCA1 Associated RING Domain (BARD1) gene are associated with breast cancer susceptibility: A case-control analysis. Breast Cancer Res. Treat. 2007;102:329–337. doi: 10.1007/s10549-006-9332-7. [DOI] [PubMed] [Google Scholar]
- 27.Sauer M.K., Andrulis I.L. Identification and characterization of missense alterations in the BRCA1 associated RING domain (BARD1) gene in breast and ovarian cancer. J. Med. Genet. 2005;42:633–638. doi: 10.1136/jmg.2004.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Brakeleer S., De Grève J., Loris R., Janin N., Lissens W., Sermijn E., Teugels E. Cancer predisposing missense and protein truncating BARD1 mutations in non-BRCA1 or BRCA2 breast cancer families. Hum. Mutat. 2010;31:E1175-85. doi: 10.1002/humu.21200. [DOI] [PubMed] [Google Scholar]
- 29.De Brakeleer S., De Grève J., Desmedt C., Joris S., Sotiriou C., Piccart M., Pauwels I., Teugels E. Frequent incidence of BARD1-truncating mutations in germline DNA from triple-negative breast cancer patients. Clin. Genet. 2016;89:336–340. doi: 10.1111/cge.12620. [DOI] [PubMed] [Google Scholar]
- 30.Toh M.R., Chong S.T., Chan S.H., Low C.E., Ishak N.D.B., Lim J.Q., Courtney E., Ngeow J. Functional analysis of clinical BARD1 germline variants. Cold Spring Harb. Mol. Case Stud. 2019;5 doi: 10.1101/mcs.a004093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber-Lassalle N., Borde J., Weber-Lassalle K., Horváth J., Niederacher D., Arnold N., Kaulfuß S., Ernst C., Paul V.G., Honisch E., et al. Germline loss-of-function variants in the BARD1 gene are associated with early-onset familial breast cancer but not ovarian cancer. Breast Cancer Res. 2019;21:55. doi: 10.1186/s13058-019-1137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ishitobi M., Miyoshi Y., Hasegawa S., Egawa C., Tamaki Y., Monden M., Noguchi S. Mutational analysis of BARD1 in familial breast cancer patients in Japan. Cancer Lett. 2003;200:1–7. doi: 10.1016/S0304-3835(03)00387-2. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez-Hormazabal P., Reyes J.M., Blanco R., Bravo T., Carrera I., Peralta O., Gomez F., Waugh E., Margarit S., Ibañez G., et al. The BARD1 Cys557Ser variant and risk of familial breast cancer in a South-American population. Mol. Biol. Rep. 2012;39:8091–8098. doi: 10.1007/s11033-012-1656-2. [DOI] [PubMed] [Google Scholar]
- 34.Gass J., Tatro M., Blackburn P., Hines S., Atwal P.S. BARD1 nonsense variant c.1921C>T in a patient with recurrent breast cancer. Clin. Case Rep. 2017;5:104–107. doi: 10.1002/ccr3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rana H.Q., Sacca R., Drogan C., Gutierrez S., Schlosnagle E., Regan M.M., Speare V., LaDuca H., Dolinsky J., Garber J.E., et al. Prevalence of germline variants in inflammatory breast cancer. Cancer. 2019;125:2194–2202. doi: 10.1002/cncr.32062. [DOI] [PubMed] [Google Scholar]
- 36.Churpek J.E., Walsh T., Zheng Y., Moton Z., Thornton A.M., Lee M.K., Casadei S., Watts A., Neistadt B., Churpek M.M., et al. Inherited predisposition to breast cancer among African American women. Breast Cancer Res. Treat. 2015;149:31–39. doi: 10.1007/s10549-014-3195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norquist B.M., Harrell M.I., Brady M.F., Walsh T., Lee M.K., Gulsuner S., Bernards S.S., Casadei S., Yi Q., Burger R.A., et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016;2:482–490. doi: 10.1001/jamaoncol.2015.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh T., Casadei S., Lee M.K., Pennil C.C., Nord A.S., Thornton A.M., Roeb W., Agnew K.J., Stray S.M., Wickramanayake A., et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc. Natl. Acad. Sci. USA. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caminsky N.G., Mucaki E.J., Perri A.M., Lu R., Knoll J.H.M., Rogan P.K. Prioritizing variants in complete hereditary breast and ovarian cancer genes in patients lacking known BRCA mutations. Hum. Mutat. 2016;37:640–652. doi: 10.1002/humu.22972. [DOI] [PubMed] [Google Scholar]
- 40.Lhotova K., Stolarova L., Zemankova P., Vocka M., Janatova M., Borecka M., Cerna M., Jelinkova S., Kral J., Volkova Z., et al. Multigene panel germline testing of 1333 czech patients with ovarian cancer. Cancers (Basel) 2020;12:956. doi: 10.3390/cancers12040956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buys S.S., Sandbach J.F., Gammon A., Patel G., Kidd J., Brown K.L., Sharma L., Saam J., Lancaster J., Daly M.B. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer. 2017;123:1721–1730. doi: 10.1002/cncr.30498. [DOI] [PubMed] [Google Scholar]
- 42.Koczkowska M., Krawczynska N., Stukan M., Kuzniacka A., Brozek I., Sniadecki M., Debniak J., Wydra D., Biernat W., Kozlowski P., et al. Spectrum and prevalence of pathogenic variants in ovarian cancer susceptibility genes in a group of 333 patients. Cancers (Basel) 2018;10:442. doi: 10.3390/cancers10110442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jian W., Shao K., Qin Q., Wang X., Song S., Wang X. Clinical and genetic characterization of hereditary breast cancer in a Chinese population. Hered. Cancer Clin. Pract. 2017;15:19. doi: 10.1186/s13053-017-0079-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pennington K.P., Walsh T., Harrell M.I., Lee M.K., Pennil C.C., Rendi M.H., Thornton A., Norquist B.M., Casadei S., Nord A.S., et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin. Cancer Res. 2014;20:764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park J.S., Lee S.-T., Nam E.J., Han J.W., Lee J.-Y., Kim J., Kim T.I., Park H.S. Variants of cancer susceptibility genes in Korean BRCA1/2 mutation-negative patients with high risk for hereditary breast cancer. BMC Cancer. 2018;18:83. doi: 10.1186/s12885-017-3940-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.González-Rivera M., Lobo M., López-Tarruella S., Jerez Y., Del Monte-Millán M., Massarrah T., Ramos-Medina R., Ocaña I., Picornell A., Santillán Garzón S., et al. Frequency of germline DNA genetic findings in an unselected prospective cohort of triple-negative breast cancer patients participating in a platinum-based neoadjuvant chemotherapy trial. Breast Cancer Res. Treat. 2016;156:507–515. doi: 10.1007/s10549-016-3792-1. [DOI] [PubMed] [Google Scholar]
- 47.Corredor J., Woodson A.H., Gutierrez Barrera A., Arun B. Multigene panel testing results in patients with multiple breast cancer primaries. Breast J. 2020;26:1337–1342. doi: 10.1111/tbj.13762. [DOI] [PubMed] [Google Scholar]
- 48.Ng P.S., Wen W.X., Fadlullah M.Z.H., Yoon S.Y., Lee S.Y., Thong M.K., Yip C.H., Mohd Taib N.A., Teo S.H. Identification of germline alterations in breast cancer predisposition genes among Malaysian breast cancer patients using panel testing. Clin. Genet. 2016;90:315–323. doi: 10.1111/cge.12735. [DOI] [PubMed] [Google Scholar]
- 49.Lolas Hamameh S., Renbaum P., Kamal L., Dweik D., Salahat M., Jaraysa T., Abu Rayyan A., Casadei S., Mandell J.B., Gulsuner S., et al. Genomic analysis of inherited breast cancer among Palestinian women: Genetic heterogeneity and a founder mutation in TP53. Int. J. Cancer. 2017;141:750–756. doi: 10.1002/ijc.30771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J., Meeks H., Feng B.-J., Healey S., Thorne H., Makunin I., Ellis J., Campbell I., Southey M., Mitchell G., et al. Targeted massively parallel sequencing of a panel of putative breast cancer susceptibility genes in a large cohort of multiple-case breast and ovarian cancer families. J. Med. Genet. 2016;53:34–42. doi: 10.1136/jmedgenet-2015-103452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crawford B., Adams S.B., Sittler T., van den Akker J., Chan S., Leitner O., Ryan L., Gil E., van ’t Veer L. Multi-Gene panel testing for hereditary cancer predisposition in unsolved high-risk breast and ovarian cancer patients. Breast Cancer Res. Treat. 2017;163:383–390. doi: 10.1007/s10549-017-4181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silva Carvalho S.D.C.E., Cury N.M., Brotto D.B., de Araujo L.F., Rosa R.C.A., Texeira L.A., Plaça J.R., Marques A.A., Peronni K.C., de Cássia Ruy P., et al. Germline variants in DNA repair genes associated with hereditary breast and ovarian cancer syndrome: Analysis of a 21 gene panel in the Brazilian population. BMC Med. Genom. 2020;13:21. doi: 10.1186/s12920-019-0652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amemiya Y., Bacopulos S., Al-Shawarby M., Al-Tamimi D., Naser W., Ahmed A., Khalifa M., Slodkowska E., Seth A. A comparative analysis of breast and ovarian cancer-related gene mutations in canadian and saudi arabian patients with breast cancer. Anticancer Res. 2015;35:2601–2610. [PubMed] [Google Scholar]
- 54.Rodríguez-Balada M., Roig B., Melé M., Albacar C., Serrano S., Salvat M., Querol M., Borràs J., Martorell L., Gumà J. Identification of germline pathogenic variants in DNA damage repair genes by a next-generation sequencing multigene panel in BRCAX patients. Clin. Biochem. 2020;76:17–23. doi: 10.1016/j.clinbiochem.2019.11.014. [DOI] [PubMed] [Google Scholar]
- 55.Slavin T.P., Maxwell K.N., Lilyquist J., Vijai J., Neuhausen S.L., Hart S.N., Ravichandran V., Thomas T., Maria A., Villano D., et al. The contribution of pathogenic variants in breast cancer susceptibility genes to familial breast cancer risk. NPJ Breast Cancer. 2017;3:22. doi: 10.1038/s41523-017-0024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castéra L., Krieger S., Rousselin A., Legros A., Baumann J.-J., Bruet O., Brault B., Fouillet R., Goardon N., Letac O., et al. Next-Generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate genes. Eur. J. Hum. Genet. 2014;22:1305–1313. doi: 10.1038/ejhg.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adedokun B., Zheng Y., Ndom P., Gakwaya A., Makumbi T., Zhou A.Y., Yoshimatsu T.F., Rodriguez A., Madduri R.K., Foster I.T., et al. Prevalence of inherited mutations in breast cancer predisposition genes among women in uganda and cameroon. Cancer Epidemiol. Biomark. Prev. 2020;29:359–367. doi: 10.1158/1055-9965.EPI-19-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li J.-Y., Jing R., Wei H., Wang M., Xiaowei Q., Liu H., Jian L., Ou J.-H., Jiang W.-H., Tian F.-G., et al. Germline mutations in 40 cancer susceptibility genes among Chinese patients with high hereditary risk breast cancer. Int. J. Cancer. 2019;144:281–289. doi: 10.1002/ijc.31601. [DOI] [PubMed] [Google Scholar]
- 59.Young E.L., Feng B.J., Stark A.W., Damiola F., Durand G., Forey N., Francy T.C., Gammon A., Kohlmann W.K., Kaphingst K.A., et al. Multigene testing of moderate-risk genes: Be mindful of the missense. J. Med. Genet. 2016;53:366–376. doi: 10.1136/jmedgenet-2015-103398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonache S., Esteban I., Moles-Fernández A., Tenés A., Duran-Lozano L., Montalban G., Bach V., Carrasco E., Gadea N., López-Fernández A., et al. Multigene panel testing beyond BRCA1/2 in breast/ovarian cancer Spanish families and clinical actionability of findings. J. Cancer Res. Clin. Oncol. 2018;144:2495–2513. doi: 10.1007/s00432-018-2763-9. [DOI] [PubMed] [Google Scholar]
- 61.Scarpitta R., Zanna I., Aretini P., Gambino G., Scatena C., Mei B., Ghilli M., Rossetti E., Roncella M., Congregati C., et al. Germline investigation in male breast cancer of DNA repair genes by next-generation sequencing. Breast Cancer Res. Treat. 2019;178:557–564. doi: 10.1007/s10549-019-05429-z. [DOI] [PubMed] [Google Scholar]
- 62.Kaur R.P., Kumar V., Shafi G., Vashistha R., Kulharia M., Munshi A. A study of mechanistic mapping of novel SNPs to male breast cancer. Med. Oncol. 2019;36:70. doi: 10.1007/s12032-019-1290-0. [DOI] [PubMed] [Google Scholar]
- 63.Arvai K.J., Roberts M.E., Torene R.I., Susswein L.R., Marshall M.L., Zhang Z., Carter N.J., Yackowski L., Rinella E.S., Klein R.T., et al. Age-Adjusted association of homologous recombination genes with ovarian cancer using clinical exomes as controls. Hered. Cancer Clin. Pract. 2019;17:19. doi: 10.1186/s13053-019-0119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cybulski C., Kluźniak W., Huzarski T., Wokołorczyk D., Kashyap A., Rusak B., Stempa K., Gronwald J., Szymiczek A., Bagherzadeh M., et al. The spectrum of mutations predisposing to familial breast cancer in Poland. Int. J. Cancer. 2019;145:3311–3320. doi: 10.1002/ijc.32492. [DOI] [PubMed] [Google Scholar]
- 65.Zeng C., Guo X., Wen W., Shi J., Long J., Cai Q., Shu X.-O., Xiang Y., Zheng W. Evaluation of pathogenetic mutations in breast cancer predisposition genes in population-based studies conducted among Chinese women. Breast Cancer Res. Treat. 2020;181:465–473. doi: 10.1007/s10549-020-05643-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shahi R.B., De Brakeleer S., Caljon B., Pauwels I., Bonduelle M., Joris S., Fontaine C., Vanhoeij M., Van Dooren S., Teugels E., et al. Identification of candidate cancer predisposing variants by performing whole-exome sequencing on index patients from BRCA1 and BRCA2-negative breast cancer families. BMC Cancer. 2019;19:313. doi: 10.1186/s12885-019-5494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Torrezan G.T., de Almeida F.G.D.S.R., Figueiredo M.C.P., de Figueiredo Barros B.D., de Paula C.A.A., Valieris R., de Souza J.E.S., Ramalho R.F., da Silva F.C.C., Ferreira E.N., et al. Complex landscape of germline variants in brazilian patients with hereditary and early onset breast cancer. Front. Genet. 2018;9:161. doi: 10.3389/fgene.2018.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rouleau E., Jesson B., Briaux A., Nogues C., Chabaud V., Demange L., Sokolowska J., Coulet F., Barouk-Simonet E., Bignon Y.J., et al. Rare germline large rearrangements in the BRCA1/2 genes and eight candidate genes in 472 patients with breast cancer predisposition. Breast Cancer Res. Treat. 2012;133:1179–1190. doi: 10.1007/s10549-012-2009-5. [DOI] [PubMed] [Google Scholar]
- 69.Esteban-Jurado C., Vila-Casadesús M., Garre P., Lozano J.J., Pristoupilova A., Beltran S., Muñoz J., Ocaña T., Balaguer F., López-Cerón M., et al. Whole-Exome sequencing identifies rare pathogenic variants in new predisposition genes for familial colorectal cancer. Genet. Med. 2015;17:131–142. doi: 10.1038/gim.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ring K.L., Bruegl A.S., Allen B.A., Elkin E.P., Singh N., Hartman A.-R., Daniels M.S., Broaddus R.R. Germline multi-gene hereditary cancer panel testing in an unselected endometrial cancer cohort. Mod. Pathol. 2016;29:1381–1389. doi: 10.1038/modpathol.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chaffee K.G., Oberg A.L., McWilliams R.R., Majithia N., Allen B.A., Kidd J., Singh N., Hartman A.-R., Wenstrup R.J., Petersen G.M. Prevalence of germ-line mutations in cancer genes among pancreatic cancer patients with a positive family history. Genet. Med. 2018;20:119–127. doi: 10.1038/gim.2017.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu C., Hart S.N., Bamlet W.R., Moore R.M., Nandakumar K., Eckloff B.W., Lee Y.K., Petersen G.M., McWilliams R.R., Couch F.J. Prevalence of pathogenic mutations in cancer predisposition genes among pancreatic cancer patients. Cancer Epidemiol. Biomark. Prev. 2016;25:207–211. doi: 10.1158/1055-9965.EPI-15-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meynard G., Mansi L., Lebahar P., Villanueva C., Klajer E., Calcagno F., Vivalta A., Chaix M., Collonge-Rame M.-A., Populaire C., et al. First description of a double heterozygosity for BRCA1 and BRCA2 pathogenic variants in a French metastatic breast cancer patient: A case report. Oncol. Rep. 2017;37:1573–1578. doi: 10.3892/or.2017.5422. [DOI] [PubMed] [Google Scholar]
- 74.Palmirotta R., Lovero D., Stucci L.S., Silvestris E., Quaresmini D., Cardascia A., Silvestris F. Double heterozygosity for BRCA1 pathogenic variant and BRCA2 polymorphic stop codon K3326X: A case report in a Southern Italian family. Int. J. Mol. Sci. 2018;19:285. doi: 10.3390/ijms19010285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ramus S.J., Friedman L.S., Gayther S.A., Ponder B.A., Bobrow L., van der Looji M., Papp J., Olah E. A breast/ovarian cancer patient with germline mutations in both BRCA1 and BRCA2. Nat. Genet. 1997;15:14–15. doi: 10.1038/ng0197-14. [DOI] [PubMed] [Google Scholar]
- 76.Lavie O., Narod S., Lejbkowicz F., Dishon S., Goldberg Y., Gemer O., Rennert G. Double heterozygosity in the BRCA1 and BRCA2 genes in the Jewish population. Ann. Oncol. 2011;22:964–966. doi: 10.1093/annonc/mdq460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Friedman E., Bar-Sade Bruchim R., Kruglikova A., Risel S., Levy-Lahad E., Halle D., Bar-On E., Gershoni-Baruch R., Dagan E., Kepten I., et al. Double heterozygotes for the Ashkenazi founder mutations in BRCA1 and BRCA2 genes. Am. J. Hum. Genet. 1998;63:1224–1227. doi: 10.1086/302040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Le Page C., Rahimi K., Rodrigues M., Heinzelmann-Schwarz V., Recio N., Tommasi S., Bataillon G., Portelance L., Golmard L., Meunier L., et al. Clinicopathological features of women with epithelial ovarian cancer and double heterozygosity for BRCA1 and BRCA2: A systematic review and case report analysis. Gynecol. Oncol. 2020;156:377–386. doi: 10.1016/j.ygyno.2019.11.019. [DOI] [PubMed] [Google Scholar]
- 79.Rebbeck T.R., Mitra N., Domchek S.M., Wan F., Chuai S., Friebel T.M., Panossian S., Spurdle A., Chenevix-Trench G., Singer C.F., et al. Modification of ovarian cancer risk by BRCA1/2-interacting genes in a multicenter cohort of BRCA1/2 mutation carriers. Cancer Res. 2009;69:5801–5810. doi: 10.1158/0008-5472.CAN-09-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spurdle A.B., Marquart L., McGuffog L., Healey S., Sinilnikova O., Wan F., Chen X., Beesley J., Singer C.F., Dressler A.-C., et al. Common genetic variation at BARD1 is not associated with breast cancer risk in BRCA1 or BRCA2 mutation carriers. Cancer Epidemiol. Biomark. Prev. 2011;20:1032–1038. doi: 10.1158/1055-9965.EPI-10-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moore J.H., Williams S.M. Epistasis and its implications for personal genetics. Am. J. Hum. Genet. 2009;85:309–320. doi: 10.1016/j.ajhg.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gibson G. Rare and common variants: Twenty arguments. Nat. Rev. Genet. 2012;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Slatkin M. Exchangeable models of complex inherited diseases. Genetics. 2008;179:2253–2261. doi: 10.1534/genetics.107.077719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Belanger M.H., Dolman L., Arcand S.L., Shen Z., Chong G., Mes-Masson A.-M., Provencher D., Tonin P.N. A targeted analysis identifies a high frequency of BRCA1 and BRCA2 mutation carriers in women with ovarian cancer from a founder population. J. Ovarian Res. 2015;8:1. doi: 10.1186/s13048-015-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Behl S., Hamel N., de Ladurantaye M., Lepage S., Lapointe R., Mes-Masson A.-M., Foulkes W.D. Founder BRCA1/BRCA2/PALB2 pathogenic variants in French-Canadian breast cancer cases and controls. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-63100-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tonin P.N., Mes-Masson A.M., Futreal P.A., Morgan K., Mahon M., Foulkes W.D., Cole D.E., Provencher D., Ghadirian P., Narod S.A. Founder BRCA1 and BRCA2 mutations in French Canadian breast and ovarian cancer families. Am. J. Hum. Genet. 1998;63:1341–1351. doi: 10.1086/302099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tonin P.N., Mes-Masson A.-M., Narod S.A., Ghadirian P., Provencher D. Founder BRCA1 and BRCA2 mutations in French Canadian ovarian cancer cases unselected for family history. Clin. Genet. 1999;55:318–324. doi: 10.1034/j.1399-0004.1999.550504.x. [DOI] [PubMed] [Google Scholar]
- 88.Tonin P.N., Perret C., Lambert J.A., Paradis A.J., Kantemiroff T., Benoît M.H., Martin G., Foulkes W.D., Ghadirian P. Founder BRCA1 and BRCA2 mutations in early-onset french Canadian breast cancer cases unselected for family history. Int. J. Cancer. 2001;95:189–193. doi: 10.1002/1097-0215(20010520)95:3<189::AID-IJC1032>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 89.Oros K.K., Ghadirian P., Greenwood C.M.T., Perret C., Shen Z., Paredes Y., Arcand S.L., Mes-Masson A.M., Narod S.A., Foulkes W.D., et al. Significant proportion of breast and/or ovarian cancer families of French Canadian descent harbor 1 of 5 BRCA1 and BRCA2 mutations. Int. J. Cancer. 2004;112:411–419. doi: 10.1002/ijc.20406. [DOI] [PubMed] [Google Scholar]
- 90.Tischkowitz M., Sabbaghian N., Hamel N., Pouchet C., Foulkes W.D., Mes-Masson A.-M., Provencher D.M., Tonin P.N. Contribution of the PALB2 c.2323C>T [p. Q775X] Founder mutation in well-defined breast and/or ovarian cancer families and unselected ovarian cancer cases of French Canadian descent. BMC Med. Genet. 2013;14:5. doi: 10.1186/1471-2350-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rivera B., Di Iorio M., Frankum J., Nadaf J., Fahiminiya S., Arcand S.L., Burk D.L., Grapton D., Tomiak E., Hastings V., et al. Functionally null RAD51D missense mutation associates strongly with ovarian carcinoma. Cancer Res. 2017;77:4517–4529. doi: 10.1158/0008-5472.CAN-17-0190. [DOI] [PubMed] [Google Scholar]
- 92.Castellsagué E., Liu J., Volenik A., Giroux S., Gagné R., Maranda B., Roussel-Jobin A., Latrielle J., Laframboise R., Palma L., et al. Characterization of a novel founder MSH6 mutation causing Lynch syndrome in the French Canadian population. Clin. Genet. 2015;87:536–542. doi: 10.1111/cge.12526. [DOI] [PubMed] [Google Scholar]
- 93.Fierheller C.T., Guitton-Sert L., Alenezi W.M., Revil T., Oros K.K., Bedard K., Arcand S.L., Serruya C., Behl S., Meunier L., et al. The genetic analysis of a founder Northern American population of European descent identifies FANCI as a candidate familial ovarian cancer risk gene. medRxiv. 2020 doi: 10.1101/2020.05.04.20090407. [DOI] [Google Scholar]
- 94.Jin Y., Xu X.L., Yang M.C., Wei F., Ayi T.C., Bowcock A.M., Baer R. Cell cycle-dependent colocalization of BARD1 and BRCA1 proteins in discrete nuclear domains. Proc. Natl. Acad. Sci. USA. 1997;94:12075–12080. doi: 10.1073/pnas.94.22.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Irminger-Finger I., Soriano J.V., Vaudan G., Montesano R., Sappino A.P. In vitro repression of Brca1-associated RING domain gene, Bard1, induces phenotypic changes in mammary epithelial cells. J. Cell Biol. 1998;143:1329–1339. doi: 10.1083/jcb.143.5.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cimmino F., Formicola D., Capasso M. Dualistic role of BARD1 in cancer. Genes (Basel) 2017;8:375. doi: 10.3390/genes8120375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bell D., Berchuck A., Birrer M., Chien J., Cramer D.W., Dao F., Dhir R., Disaia P., Gabra H., Glenn P., et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166.Integrated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koboldt D.C., Fulton R.S., McLellan M.D., Schmidt H., Kalicki-Veizer J., McMichael J.F., Fulton L.L., Dooling D.J., Ding L., Mardis E.R., et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zack T.I., Schumacher S.E., Carter S.L., Cherniack A.D., Saksena G., Tabak B., Lawrence M.S., Zhang C.Z., Wala J., Mermel C.H., et al. Pan-Cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Campbell P.J., Getz G., Korbel J.O., Stuart J.M., Jennings J.L., Stein L.D., Perry M.D., Nahal-Bose H.K., Ouellette B.F.F., Li C.H., et al. Pan-Cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McCarthy E.E., Celebi J.T., Baer R., Ludwig T. Loss of Bard1, the heterodimeric partner of the brca1 tumor suppressor, results in early embryonic lethality and chromosomal instability. Mol. Cell. Biol. 2003;23:5056–5063. doi: 10.1128/MCB.23.14.5056-5063.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shakya R., Szabolcs M., McCarthy E., Ospina E., Basso K., Nandula S., Murty V., Baer R., Ludwig T. The basal-like mammary carcinomas induced by Brca1 or Bard1 inactivation implicate the BRCA1/BARD1 heterodimer in tumor suppression. Proc. Natl. Acad. Sci. USA. 2008;105:7040–7045. doi: 10.1073/pnas.0711032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ayi T.C., Tsan J.T., Hwang L.Y., Bowcock A.M., Baer R. Conservation of function and primary structure in the BRCA1-associated RING domain (BARD1) protein. Oncogene. 1998;17:2143–2148. doi: 10.1038/sj.onc.1202123. [DOI] [PubMed] [Google Scholar]
- 104.Fox D., III, Le Trong I., Rajagopal P., Brzovic P.S., Stenkamp R.E., Klevit R.E. Crystal structure of the BARD1 ankyrin repeat domain and its functional consequences. J. Biol. Chem. 2008;283:21179–21186. doi: 10.1074/jbc.M802333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tarsounas M., Sung P. The antitumorigenic roles of BRCA1–BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol. 2020;21:284–299. doi: 10.1038/s41580-020-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meza J.E., Brzovic P.S., King M.C., Klevit R.E. Mapping the functional domains of BRCA1. Interaction of the ring finger domains of BRCA1 and BARD1. J. Biol. Chem. 1999;274:5659–5665. doi: 10.1074/jbc.274.9.5659. [DOI] [PubMed] [Google Scholar]
- 107.Brzovic P.S., Rajagopal P., Hoyt D.W., King M.C., Klevit R.E. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 108.Joukov V., Chen J., Fox E.A., Green J.B., Livingston D.M. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl. Acad. Sci. USA. 2001;98:12078–12083. doi: 10.1073/pnas.211427098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Choudhury A.D., Xu H., Baer R. Ubiquitination and proteasomal degradation of the BRCA1 tumor suppressor is regulated during cell cycle progression. J. Biol. Chem. 2004;279:33909–33918. doi: 10.1074/jbc.M403646200. [DOI] [PubMed] [Google Scholar]
- 110.Lorick K.L., Jensen J.P., Fang S., Ong A.M., Hatakeyama S., Weissman A.M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Buetow L., Huang D.T. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2016;17:626–642. doi: 10.1038/nrm.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Senft D., Qi J., Ronai Z.A. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer. 2018;18:69–88. doi: 10.1038/nrc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hashizume R., Fukuda M., Maeda I., Nishikawa H., Oyake D., Yabuki Y., Ogata H., Ohta T. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- 114.Ruffner H., Joazeiro C.A.P., Hemmati D., Hunter T., Verma I.M. Cancer-Predisposing mutations within the RING domain of BRCA1: Loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl. Acad. Sci. USA. 2001;98:5134–5139. doi: 10.1073/pnas.081068398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brzovic P.S., Keeffe J.R., Nishikawa H., Miyamoto K., Fox D., III, Fukuda M., Ohta T., Klevit R. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. USA. 2003;100:5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brzovic P.S., Meza J.E., King M.C., Klevit R.E. BRCA1 RING domain cancer-predisposing mutations. Structural consequences and effects on protein-protein interactions. J. Biol. Chem. 2001;276:41399–41406. doi: 10.1074/jbc.M106551200. [DOI] [PubMed] [Google Scholar]
- 117.Stewart M.D., Zelin E., Dhall A., Walsh T., Upadhyay E., Corn J.E., Chatterjee C., King M.-C., Klevit R.E. BARD1 is necessary for ubiquitylation of nucleosomal histone H2A and for transcriptional regulation of estrogen metabolism genes. Proc. Natl. Acad. Sci. USA. 2018;115:1316–1321. doi: 10.1073/pnas.1715467115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Adamovich A.I., Banerjee T., Wingo M., Duncan K., Ning J., Martins Rodrigues F., Huang K.-L., Lee C., Chen F., Ding L., et al. Functional analysis of BARD1 missense variants in homology-directed repair and damage sensitivity. PLoS Genet. 2019;15:e1008049. doi: 10.1371/journal.pgen.1008049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Irminger-Finger I., Jefford C.E. Is there more to BARD1 than BRCA1? Nat. Rev. Cancer. 2006;6:382–391. doi: 10.1038/nrc1878. [DOI] [PubMed] [Google Scholar]
- 120.Nakamura K., Saredi G., Becker J.R., Foster B.M., Nguyen N.V., Beyer T.E., Cesa L.C., Faull P.A., Lukauskas S., Frimurer T., et al. H4K20me0 recognition by BRCA1–BARD1 directs homologous recombination to sister chromatids. Nat. Cell Biol. 2019;21:311–318. doi: 10.1038/s41556-019-0282-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tsuzuki M., Wu W., Nishikawa H., Hayami R., Oyake D., Yabuki Y., Fukuda M., Ohta T. A truncated splice variant of human BARD1 that lacks the RING finger and ankyrin repeats. Cancer Lett. 2006;233:108–116. doi: 10.1016/j.canlet.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 122.Ratajska M., Matusiak M., Kuzniacka A., Wasag B., Brozek I., Biernat W., Koczkowska M., Debniak J., Sniadecki M., Kozlowski P., et al. Cancer predisposing BARD1 mutations affect exon skipping and are associated with overexpression of specific BARD1 isoforms. Oncol. Rep. 2015;34:2609–2617. doi: 10.3892/or.2015.4235. [DOI] [PubMed] [Google Scholar]
- 123.Billing D., Horiguchi M., Wu-Baer F., Taglialatela A., Leuzzi G., Nanez S.A., Jiang W., Zha S., Szabolcs M., Lin C.-S., et al. The BRCT domains of the BRCA1 and BARD1 tumor suppressors differentially regulate homology-directed repair and stalled fork protection. Mol. Cell. 2018;72:127–139.e8. doi: 10.1016/j.molcel.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li M., Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013;23:693–704. doi: 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Choudhary R.K., Siddiqui M.Q., Thapa P.S., Gadewal N., Nachimuthu S.K., Varma A.K. Structural basis to stabilize the domain motion of BARD1-ARD BRCT by CstF50. Sci. Rep. 2017;7:3849. doi: 10.1038/s41598-017-03816-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Laufer M., Nandula S.V., Modi A.P., Wang S., Jasin M., Murty V.V.V.S., Ludwig T., Baer R. Structural requirements for the BARD1 tumor suppressor in chromosomal stability and homology-directed DNA repair. J. Biol. Chem. 2007;282:34325–34333. doi: 10.1074/jbc.M705198200. [DOI] [PubMed] [Google Scholar]
- 127.Irminger-Finger I., Leung W.-C., Li J., Dubois-Dauphin M., Harb J., Feki A., Jefford C.E., Soriano J.V., Jaconi M., Montesano R., et al. Identification of BARD1 as Mediator between proapoptotic stress and p53-Dependent apoptosis. Mol. Cell. 2001;8:1255–1266. doi: 10.1016/S1097-2765(01)00406-3. [DOI] [PubMed] [Google Scholar]
- 128.Kleiman F.E., Manley J.L. The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell. 2001;104:743–753. doi: 10.1016/S0092-8674(01)00270-7. [DOI] [PubMed] [Google Scholar]
- 129.Lee C., Banerjee T., Gillespie J., Ceravolo A., Parvinsmith M.R., Starita L.M., Fields S., Toland A.E., Parvin J.D. Functional analysis of BARD1 missense variants in homology-directed repair of DNA double strand breaks. Hum. Mutat. 2015;36:1205–1214. doi: 10.1002/humu.22902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Casadei S., Gulsuner S., Shirts B.H., Mandell J.B., Kortbawi H.M., Norquist B.S., Swisher E.M., Lee M.K., Goldberg Y., O’Connor R., et al. Characterization of splice-altering mutations in inherited predisposition to cancer. Proc. Natl. Acad. Sci. USA. 2019;116:26798–26807. doi: 10.1073/pnas.1915608116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yoshino Y., Qi H., Fujita H., Shirota M., Abe S., Komiyama Y., Shindo K., Nakayama M., Matsuzawa A., Kobayashi A., et al. BRCA1-Interacting Protein OLA1 requires interaction with BARD1 to regulate centrosome number. Mol. Cancer Res. 2018;16:1499–1511. doi: 10.1158/1541-7786.MCR-18-0269. [DOI] [PubMed] [Google Scholar]
- 132.Chen H., Cohen P., Chen S. How big is a big odds ratio? Interpreting the magnitudes of odds ratios in epidemiological studies. Commun. Stat. Simul. Comput. 2010;39:860–864. doi: 10.1080/03610911003650383. [DOI] [Google Scholar]
- 133.Liu J., Cristea M.C., Frankel P., Neuhausen S.L., Steele L., Engelstaedter V., Matulonis U., Sand S., Tung N., Garber J.E., et al. Clinical characteristics and outcomes of BRCA-associated ovarian cancer: Genotype and survival. Cancer Genet. 2012;205:34–41. doi: 10.1016/j.cancergen.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Suszynska M., Klonowska K., Jasinska A.J., Kozlowski P. Large-Scale meta-analysis of mutations identified in panels of breast/ovarian cancer-related genes—Providing evidence of cancer predisposition genes. Gynecol. Oncol. 2019;153:452–462. doi: 10.1016/j.ygyno.2019.01.027. [DOI] [PubMed] [Google Scholar]
- 135.Ding D.-P., Zhang Y., Ma W.-L., He X.-F., Wang W., Yu H.-L., Guo Y.-B., Zheng W.-L. Lack of association between BARD1 Cys557Ser variant and breast cancer risk: A meta-analysis of 11,870 cases and 7,687 controls. J. Cancer Res. Clin. Oncol. 2011;137:1463–1468. doi: 10.1007/s00432-011-1006-0. [DOI] [PubMed] [Google Scholar]
- 136.Tung N.M., Boughey J.C., Pierce L.J., Robson M.E., Bedrosian I., Dietz J.R., Dragun A., Gelpi J.B., Hofstatter E.W., Isaacs C.J., et al. Management of hereditary breast cancer: American society of clinical oncology, American society for radiation oncology, and society of surgical oncology guideline. J. Clin. Oncol. 2020;38:2080–2106. doi: 10.1200/JCO.20.00299. [DOI] [PubMed] [Google Scholar]
- 137.Pal T., Agnese D., Daly M., La Spada A., Litton J., Wick M., Klugman S., Esplin E.D., Jarvik G.P. Points to consider: Is there evidence to support BRCA1/2 and other inherited breast cancer genetic testing for all breast cancer patients? A statement of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2020;22:681–685. doi: 10.1038/s41436-019-0712-x. [DOI] [PubMed] [Google Scholar]
- 138.Plon S.E., Eccles D.M., Easton D., Foulkes W.D., Genuardi M., Greenblatt M.S., Hogervorst F.B.L., Hoogerbrugge N., Spurdle A.B., Tavtigian S.V. Sequence variant classification and reporting: Recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 2008;29:1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Konstantinopoulos P.A., Norquist B., Lacchetti C., Armstrong D., Grisham R.N., Goodfellow P.J., Kohn E.C., Levine D.A., Liu J.F., Lu K.H., et al. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline. J. Clin. Oncol. 2020;38:1222–1245. doi: 10.1200/JCO.19.02960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:1062–1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cline M.S., Liao R.G., Parsons M.T., Paten B., Alquaddoomi F., Antoniou A., Baxter S., Brody L., Cook-Deegan R., Coffin A., et al. BRCA Challenge: BRCA Exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genet. 2018;14:e1007752. doi: 10.1371/journal.pgen.1007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hart S.N., Polley E.C., Yussuf A., Yadav S., Goldgar D.E., Hu C., LaDuca H., Smith L.P., Fujimoto J., Li S., et al. Mutation prevalence tables for hereditary cancer derived from multi-gene panel testing. Hum. Mutat. 2020;92:220–225. doi: 10.1002/humu.24053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.PDQ Cancer Genetics Editorial Board . Genetics of Breast and Gynecologic Cancers (PDQ®): Health Professional Version. National Cancer Institute (US); Bethesda, MD, USA: 2020. [PubMed] [Google Scholar]
- 145.Yang X., Song H., Leslie G., Engel C., Hahnen E., Auber B., Horváth J., Kast K., Niederacher D., Turnbull C., et al. Ovarian and breast cancer risks associated with pathogenic variants in RAD51C and RAD51D. J. Natl. Cancer Inst. 2020:djaa030. doi: 10.1093/jnci/djaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yang X., Leslie G., Doroszuk A., Schneider S., Allen J., Decker B., Dunning A.M., Redman J., Scarth J., Plaskocinska I., et al. Cancer risks associated with germline PALB2 pathogenic variants: An international study of 524 families. J. Clin. Oncol. 2020;38:674–685. doi: 10.1200/JCO.19.01907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Meeks H.D., Song H., Michailidou K., Bolla M.K., Dennis J., Wang Q., Barrowdale D., Frost D., McGuffog L., Ellis S., et al. BRCA2 Polymorphic Stop Codon K3326X and the risk of breast, prostate, and ovarian cancers. J. Natl. Cancer Inst. 2016;108 doi: 10.1093/jnci/djv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Peterlongo P., Catucci I., Colombo M., Caleca L., Mucaki E., Bogliolo M., Marin M., Damiola F., Bernard L., Pensotti V., et al. FANCM c.5791C>T nonsense mutation (rs144567652) induces exon skipping, affects DNA repair activity and is a familial breast cancer risk factor. Hum. Mol. Genet. 2015;24:5345–5355. doi: 10.1093/hmg/ddv251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Arem H., Loftfield E. Cancer epidemiology: A survey of modifiable risk factors for prevention and survivorship. Am. J. Lifestyle Med. 2018;12:200–210. doi: 10.1177/1559827617700600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Moorman P.G., Havrilesky L.J., Gierisch J.M., Coeytaux R.R., Lowery W.J., Urrutia R.P., Dinan M., Mcbroom A.J., Hasselblad V., Sanders G.D., et al. Oral contraceptives and risk of ovarian cancer and breast cancer among high-risk women: A systematic review and meta-analysis. J. Clin. Oncol. 2013;31:4188–4198. doi: 10.1200/JCO.2013.48.9021. [DOI] [PubMed] [Google Scholar]
- 151.Milne R.L., Osorio A., Ramón y Cajal T., Baiget M., Lasa A., Diaz-Rubio E., de la Hoya M., Caldés T., Teulé A., Lázaro C., et al. Parity and the risk of breast and ovarian cancer in BRCA1 and BRCA2 mutation carriers. Breast Cancer Res. Treat. 2010;119:221–232. doi: 10.1007/s10549-009-0394-1. [DOI] [PubMed] [Google Scholar]
- 152.Hartmann L.C., Sellers T.A., Schaid D.J., Frank T.S., Soderberg C.L., Sitta D.L., Frost M.H., Grant C.S., Donohue J.H., Woods J.E., et al. Efficacy of bilateral prophylactic mastectomy in BRCA1 and BRCA2 gene mutation carriers. J. Natl. Cancer Inst. 2001;93:1633–1637. doi: 10.1093/jnci/93.21.1633. [DOI] [PubMed] [Google Scholar]
- 153.Rebbeck T.R., Lynch H.T., Neuhausen S.L., Narod S.A., van’t Veer L., Garber J.E., Evans G., Isaacs C., Daly M.B., Matloff E., et al. Prophylactic oophorectomy in carriers of BRCA1 or BRCA2 mutations. N. Engl. J. Med. 2002;346:1616–1622. doi: 10.1056/NEJMoa012158. [DOI] [PubMed] [Google Scholar]
- 154.Meijers-Heijboer H., van Geel B., van Putten W.L., Henzen-Logmans S.C., Seynaeve C., Menke-Pluymers M.B., Bartels C.C., Verhoog L.C., van den Ouweland A.M., Niermeijer M.F., et al. Breast cancer after prophylactic bilateral mastectomy in women with a BRCA1 or BRCA2 mutation. N. Engl. J. Med. 2001;345:159–164. doi: 10.1056/NEJM200107193450301. [DOI] [PubMed] [Google Scholar]
- 155.Bolton K.L., Chenevix-Trench G., Goh C., Sadetzki S., Ramus S.J., Karlan B.Y., Lambrechts D., Despierre E., Barrowdale D., McGuffog L., et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012;307:382–390. doi: 10.1001/jama.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mylavarapu S., Das A., Roy M. Role of BRCA mutations in the modulation of response to platinum therapy. Front. Oncol. 2018;8:16. doi: 10.3389/fonc.2018.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fong P.C., Yap T.A., Boss D.S., Carden C.P., Mergui-Roelvink M., Gourley C., De Greve J., Lubinski J., Shanley S., Messiou C., et al. Poly (ADP)-Ribose polymerase inhibition: Frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J. Clin. Oncol. 2010;28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 158.Audeh M.W., Carmichael J., Penson R.T., Friedlander M., Powell B., Bell-Mcguinn K.M., Scott C., Weitzel J.N., Oaknin A., Loman N., et al. Oral poly (ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet Oncol. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 159.Gelmon K.A., Tischkowitz M., Mackay H., Swenerton K., Robidoux A., Tonkin K., Hirte H., Huntsman D., Clemons M., Gilks B., et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: A phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- 160.Kaufman B., Shapira-Frommer R., Schmutzler R.K., Audeh M.W., Friedlander M., Balmana J., Mitchell G., Fried G., Stemmer S.M., Hubert A., et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. 2015;33:244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pujade-Lauraine E., Ledermann J.A., Selle F., Gebski V., Penson R.T., Oza A.M., Korach J., Huzarski T., Poveda A., Pignata S., et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18:1274–1284. doi: 10.1016/S1470-2045(17)30469-2. [DOI] [PubMed] [Google Scholar]
- 162.Ledermann J., Harter P., Gourley C., Friedlander M., Vergote I., Rustin G., Scott C.L., Meier W., Shapira-Frommer R., Safra T., et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: A preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.