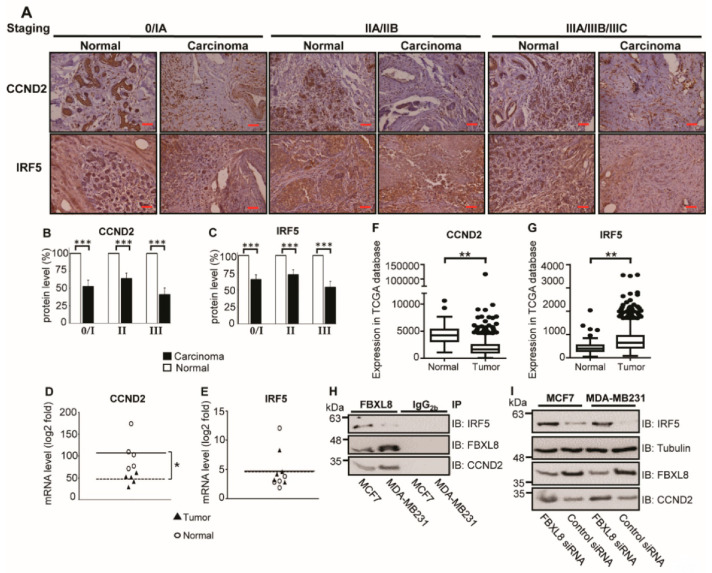
Figure 6.
FBXL8 pulled down CCND2 and IRF5, and knockdown of FBXL8 accumulated CCND2 and IRF5 protein levels in BRCA cells. To clarify the potential of CCND2 and IRF5 in primary BRCA tissues, retrospective analyses were performed by IHC (A–C) and NGS-based RNA-Seq (D,E), and from TCGA database (F,G). (A) immunofluorescence staining of CCND2 and IRF5. Protein expression levels of CCND2 and IRF5 are examined in both normal (n = 30) and carcinoma (n = 30) breast tissues. The corresponding quantitative results of CCND2 and IRF5 in IHC are shown in panels (B,C), respectively. (D,E) Quantitative results from each patient. Each data point represents a log2 fold change value for CCND2 and IRF5 (*, FDR < 0.05). (F,G) Box-whisker plot showing the transcript levels of CCND2 and IRF5 in TCGA database, respectively. Vertical lines, box and horizontal white line correspond to min-max range, 25–75th percentile range and median, respectively. Data are reported as normalized count as provided in the TCGA level 3 data (**, p < 0.01). The corresponding clinicopathological information for IHC are shown in Figure S2. Brown color indicates DAB dye-stained protein of interest. Scale bar is 100 μm, shown as the red color line (—). (H) Co-IP of of FBXL8 and CCND2 (or IRF5) in BRCA cells; total cell lysates were immunoprecipitated with anti-FBXL8 (or control anti-IgG2b) antibody, separated by 10% SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis), immunoblotted and probed with FBXL8, CCND2 or IRF5 antibodies. (I) To confirm whether CCND2 and IRF5 are regulated by FBXL8-dependent protein degradation, FBXL8 RNAi was performed in MCF7 and MDA-MB231 cells, followed by immunoblotting analysis. Tubulin was used as a loading control for immunoblotting.