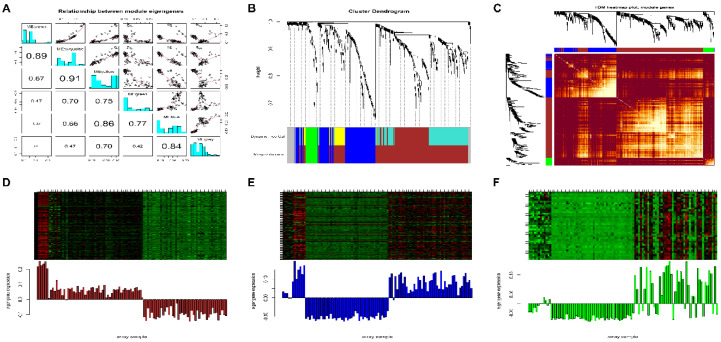
Figure 2.
(A) Pairwise scatter plots among the MEs of original modules and samples (arrays). Each dot signifies a microarray sample. MEbrown, MEturquoise, MEyellow, MEgreen, MEblue, and MEgrey denote the ME of brown, turquoise, yellow, green, blue, and grey modules, respectively. Absolute values of corresponding correlations are denoted by the numbers below the diagonal. Frequency plots (histograms) of the variables are plotted along the diagonal. Yellow and turquoise modules are highly correlated (i.e., r = 0.91) followed by brown and turquoise modules (i.e., r = 0.89). Both turquoise and yellow modules were merged to the brown module. (B) Hierarchical clustering dendrogram of DEGs clustered based on a dissimilarity measure (1-TOM) together with original module colors (6) and merged module colors (4). The four merged modules (or meta-modules) contained highly co-expressed genes with size as follows: brown (361 DEGs), blue (163 DEGs), green (39 DEGs), and grey (40 DEGs) respectively. (C) Topological overlap matrix (TOM) heatmap plot where the rows and columns correspond to single genes. Lighter color signifies low topological overlap and progressively darker orange and red colors signify higher topological overlap. Dark colored blocks along the diagonal signify the meta-modules. The corresponding gene dendrogram and module assignment are also displayed along the left and the top sides of the plot. Expression heatmap of (D) brown, (E) blue, and (F) green meta-module genes where the rows and columns correspond to genes and samples, respectively. The green and red colored bands in the heatmap denote lower and higher expression level of genes for each module. Also, the corresponding module eigengene expression levels (y-axis) across the same samples (x-axis) are displayed at the bottom panel of each module heatmap in the form of barplot. The module eigengene levels are directly correlated with the gene expression levels of each corresponding module.