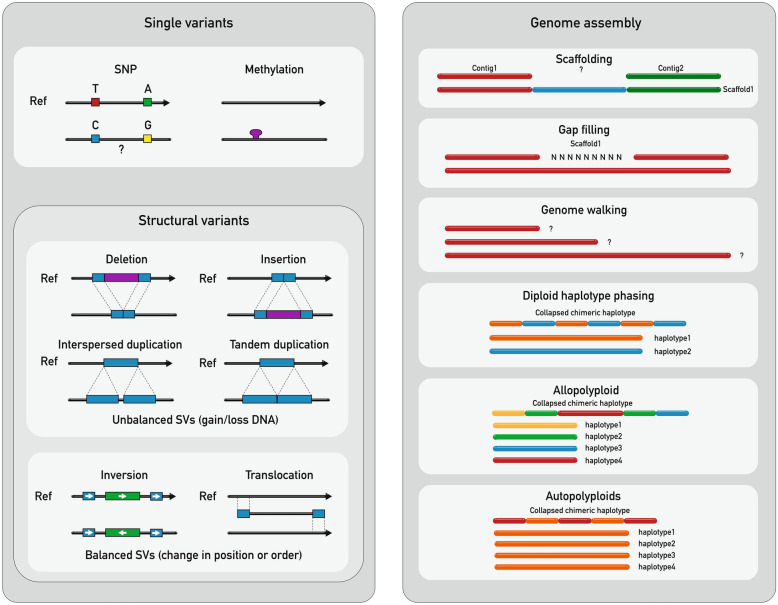
Fig. 5.
CRISPR-Cas9 targeted enrichment applications. The method can be used to identify single nucleotide genomic and epigenomic variants as well as long structural variants (SVs) (both, balanced and unbalanced SVs) by knowing one or both ends of the region of interest (ROI). It can improve genome assembly contiguity by assisting in the scaffolding and gap filling processes when two ends of the ROI are known or by genome walking approaches when just one side of the ROI is known. Phasing targeted loci of genome assemblies for both diploids and polyploid species is also possible with this technique