Figure 5.
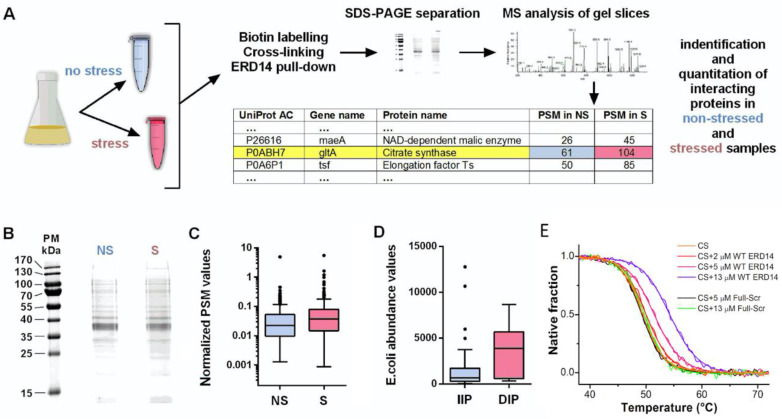
Identification of potential interaction partners of ERD14. (A) Brief overview of the identification of the binding partners of ERD14 in the cell: biotinylated ERD14 was overexpressed and cells were treated with a membrane-permeable cross-linker, either without or with prior exposure to heat stress (50 °C × 15 min). Protein partners bound to ERD14 were affinity purified with Streptavidin–Sepharose, separated on SDS-PAGE, in-gel digested by trypsin, and analyzed by LC-MS/MS (for details, cf. Materials and Methods, for data, cf. Table S3 (Supplementary Materials)). (B) SDS-PAGE of cross-linked proteins without (NS) and with (S) stress treatment. (C) The distribution of normalized copy numbers of proteins (peptide spectrum matches, or PSM) cross-linked to ERD14 before and after heat stress shows an increased number of proteins interacting with ERD14 after stress (p = 0.004). (D) Average cellular abundances of the identified proteins with increased (IIP) or decreased (DIP) ERD14 interaction potential are significantly different (p = 0.0002), showing that increased binding to ERD14 upon stress is not due to protein abundances. (E) Thermal denaturation of citrate synthase in the presence of ERD14 reveals their interaction. First, 2 μM citrate synthase (CS) was heated up in the presence of various concentrations of WT ERD14 (0—orange, 2—red, 5—magenta and 13 μM—purple) or its scrambled variant (5—black and 13 μM—green). Thermal denaturation was followed by the CD signal at 220 nm. The presence of WT ERD14 increased the melting point of CS and altered the sharpness of the unfolding transition in a concentration-dependent manner. Despite its similar physicochemical properties, the Full-Scr ERD14 variant had no relevant effect on the thermal denaturation of CS. Normalized thermograms are shown. Data was fitted according to Shih et al. [40].