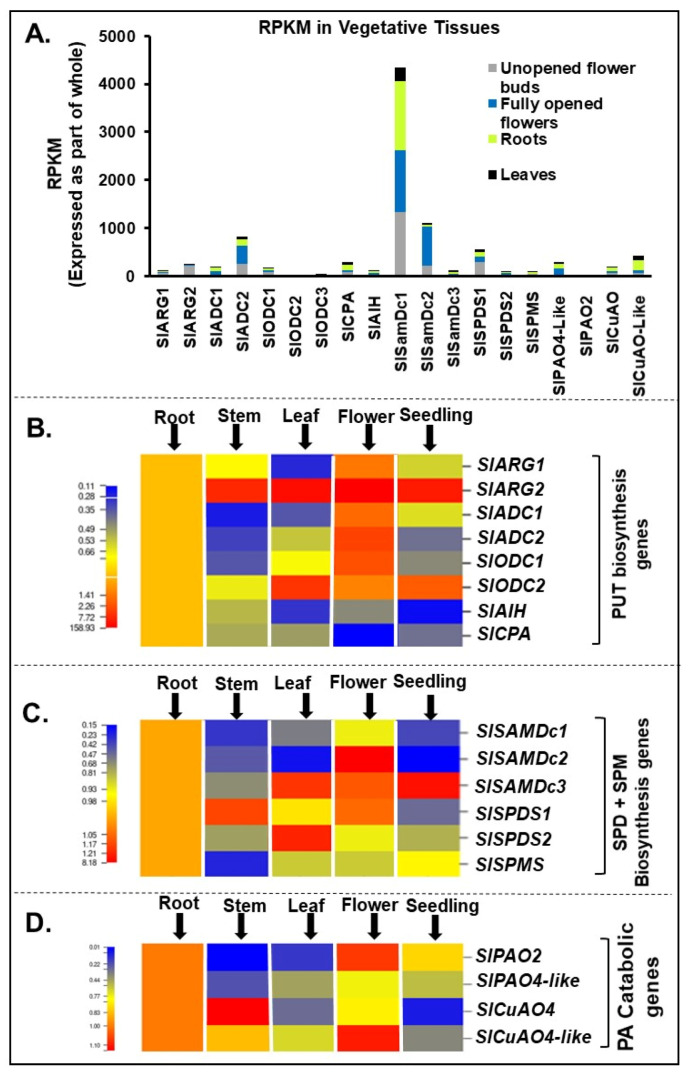
Figure 1.
Spatiotemporal gene expression of tomato (Solanum lycopersicum L.) polyamines (PA) metabolic pathway genes. (A) Global transcriptome analysis of tomato vegetative tissues (flower-buds, open flower, leaf and roots) reveals differential abundance of polyamine biosynthesis genes SlARG1, 2, SlADC1, 2 SlODC1, 2, SlAIH and SlCPA and catabolic enzymes encoding genes SlPAO2 and SlPAO4-Like; SlCuAO and SlCuAO4-like. (B) qRT-PCR based gene expression analysis of tomato vegetative tissues (root, stem, leaf, open flower and seedling tissues) reveals differential abundance of putrescine (PUT) biosynthesis genes SlARG1, 2, SlADC1, 2 SlODC1, 2, SlAIH and SlCPA. (C) Spermidine (SPD) and spermine (SPM) biosynthesis genes SlSAMDc1, 2, 3, SlSPDS1, 2 and SlSPMS. (D) PA catabolic pathway genes SlPAO2 and SlPAO4-Like; SlCuAO and SlCuAO4-like. A minimum of three plants were taken for collection of vegetative tissues for each biological replicate and mean data points and standard error were derived from three biological replicates. Statistical significance (p-value) was derived using Graph prism pad program via Tukey test/t-test. SlUBI3 and SlTIP41 were used as reference genes.