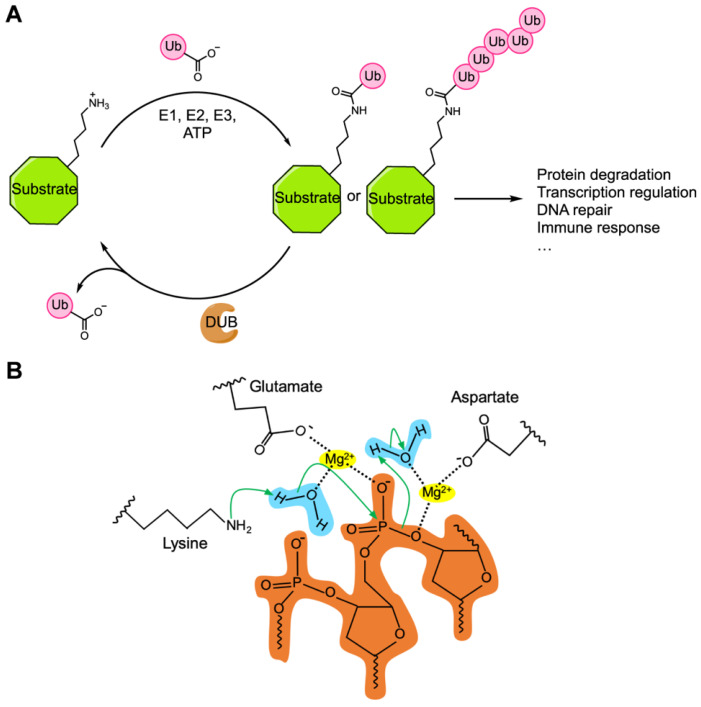
Figure 2.
(A) Summary of the ubiquitin system. In eukaryotes, the small protein modifier ubiquitin (Ub) can be covalently attached to a nucleophilic residue of its protein substrate by the combined actions of the E1 ubiquitin-activating enzyme, an E2 ubiquitin conjugating enzyme, and an E3 ubiquitin ligase in an ATP-dependent manner. Polyubiquitin chains can form when a ubiquitin is further modified by other ubiquitin molecules. Ubiquitin modification is reversed by the enzymatic activity of deubiquitylases (DUBs). (B) Mechanism of PD-(D/E)xK nucleases. Catalytic aspartate and glutamate coordinate two magnesium ions (yellow) that bring two water molecules (blue) near the cleavage site. Catalytic lysine deprotonates and activates one of the water molecules, which then serves as a nucleophile that attacks the DNA phosphate backbone (orange). The second water molecule, along with the two magnesium ions, help stabilize the reaction intermediate. Green arrows represent electron pair movements.