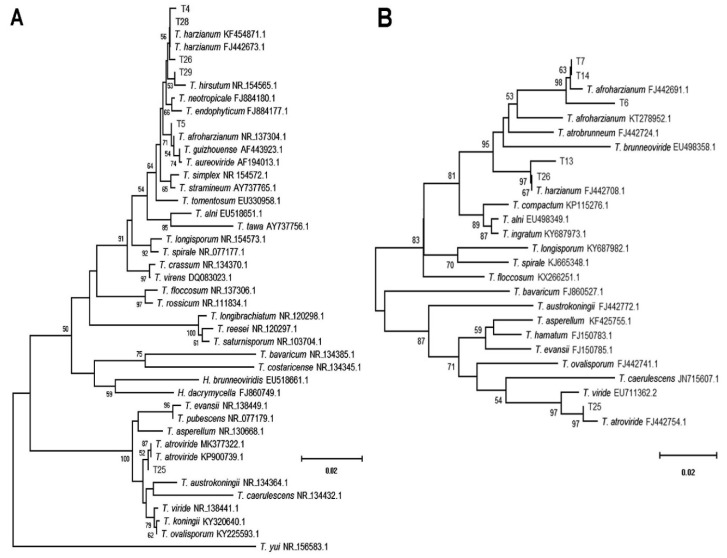
Figure 1.
The phylogenetic distance trees of Trichoderma spp. isolates with the closest species (including Hypocrea spp. anamorphs), based on sequenced PCR products and available GenBank accessions. Dendrograms inferred with MEGA X, applying the neighbor-joining and Jukes–Cantor methods to the aligned sequences of the ITS1-2 (A) and Rpb2 (B) gene regions. The percent of trees in which the associated taxa clustered together in the bootstrap test (100 replicates) are shown next to the branches (threshold: 0.5). Optimal trees, with the sum of the branch lengths = 0.54914559 (A) and 0.56187331 (B), are drawn to scale, in the number of base substitutions per site.