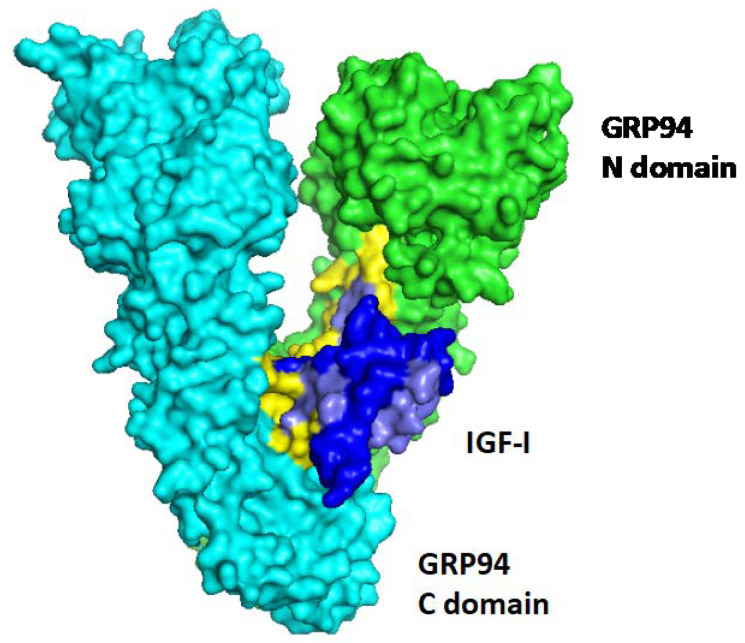
Figure 1.
A predicted complex between GRP94 and IGF-I. The crystal structure of human IGF-1 (1IMX [63]) was docked onto the crystal structure of GRP94 (2O1U [64]) with the ZDOCK algorithm (version 3.0.2 [65]). The two monomers of GRP94, are shown in cyan and green, with the N-terminal and C-terminal domains indicated. The interacting amino acids are colored yellow. The complex shown is the highest scoring predicted complex, and eight other complexes out of the 10 highest scoring ones overlap with it, predicting the same topology of binding. The GRP94 interacting residues are from from the internal face of the Middle domain and the C-terminal domain of the chaperone. The IGF-I interacting residues are mostly derived from its N-terminal 32 amino acids, colored light blue, while the C-terminal 28 residues (deep blue) are mostly predicted as non-interacting.