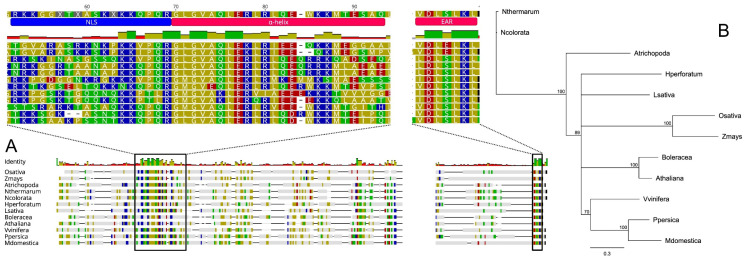
Figure 2.
Bioinformatic analysis of the SPL/NZZ gene homologs in plant species representing the two major groups of angiosperms, monocotyledons and dicotyledons, including different basal angiosperm lineages belonging to the families Amborellaceae and Nymphaeaceae. (A) MUSCLE-based amino acid alignment of 11 putative SPL/NOZZLE sequences retrieved from Zea mays (KY110964.1), Oryza sativa (LOC_Os01g11430.1), Hypericum perforatum (apomictic species, OBUPD-D1 Hpctg51499), Brassica oleracea (Bol013057), Malus domestica (MD11G1234600), Prunus persica (Prupe.4G192500.1), Vitis vinifera (VIT_219s0014g03940.1), Lactuca sativa (Lsat_1_v5_gn_0_5400), Amborella trichopoda (XP_006833114.1), Nymphaea colorata (XP031473161.1), Nymphaea thermarum (KAF3782288.1), putative orthologues of SPL protein of Arabidopsis thaliana (AT4G27330). On the top of the panel the three main functional domains that resulted conserved among the 12 proteins are highlighted. NLS is a basic region rich in Arginine (R) and Lysine (K) that is thought to represent a putative nuclear localization signal; the α-helix sequence (also known as SPL-motif) is crucial for the constitution of homodimers and heterodimers, binding and inhibiting CINCINNATA (CIN)-like TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors (TF); the EAR motif in the C-terminal region recruits TOPLESS/TOPLESS-RELATED (TPL/TPR) proteins to co-suppress the activity of the CIN-like TCP family. (B) Similarity-based neighbor-joining analysis performed using the 12 amino acid sequences with bootstrap values supporting all major nodes.