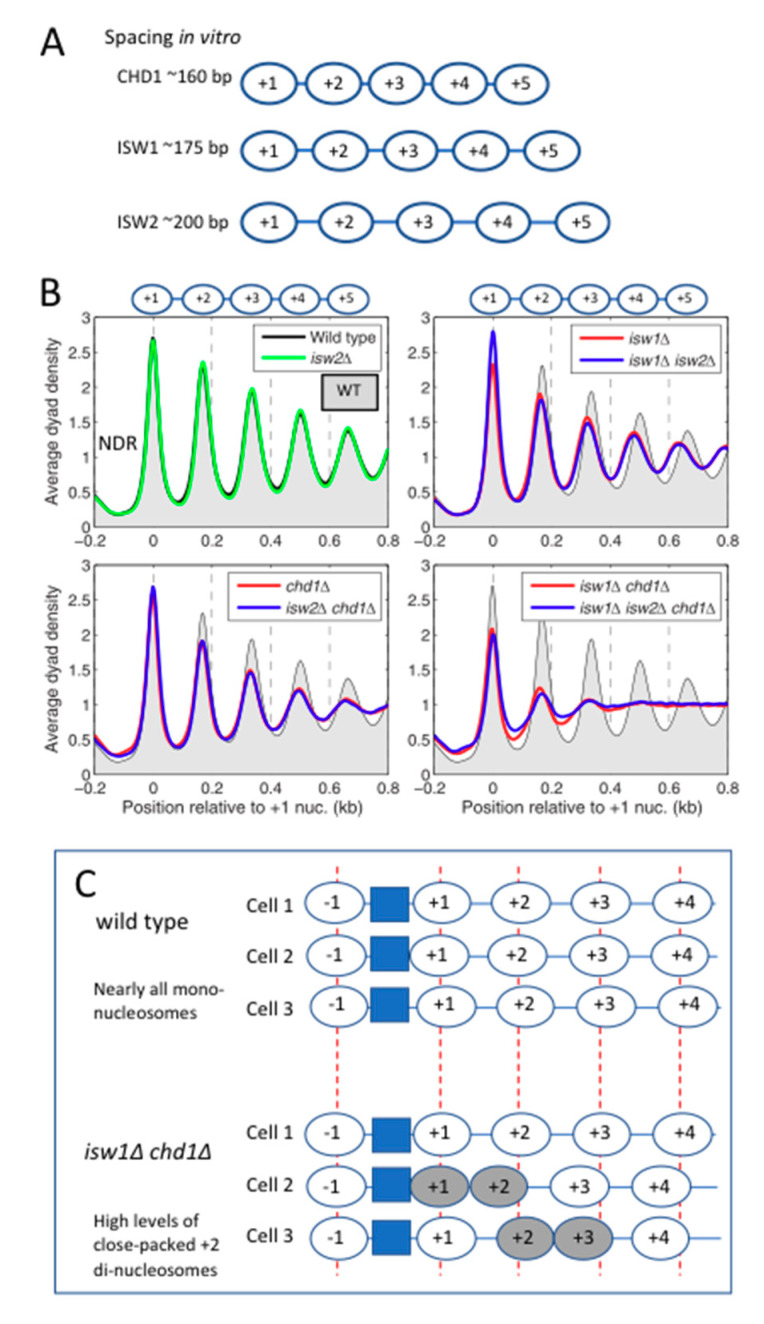
Figure 4.
Yeast nucleosome-spacing enzymes in vitro and in vivo. (A) CHD1, ISW1 and ISW2 space nucleosomes differently in vitro. The nucleosomes are not phased. (B) The effects of null mutations in CHD1, ISW1 and ISW2 on chromatin organisation. Dyad phasing in wild type (WT) cells (black line with grey fill). All ~5700 yeast genes are aligned on the first (+1) nucleosome (set at 0). The genomic average dyad density is set at 1. The promoters are nucleosome-depleted regions (NDRs). Loss of ISW2 has no effect. Spacing is reduced in isw1Δ cells, while phasing is weaker in both isw1Δ and chd1Δ cells. Chromatin organisation in the isw1Δ chd1Δ double mutant is severely disrupted. A mathematical method to quantify the degree of phasing based on the relative height and width of each nucleosome peak is available [73]. Adapted from [73]. (C) Proposed contribution of close-packed di-nucleosomes involving the +1/+2 or +2/+3 nucleosomes (grey ovals) to phasing disruption in the isw1Δ chd1Δ double mutant. A di-nucleosome will alter the positions of downstream nucleosomes. In addition, the nucleosomes included in di-nucleosomes will be missing from this mono-nucleosome analysis, resulting in an apparent decrease in signal around the +2 nucleosome.