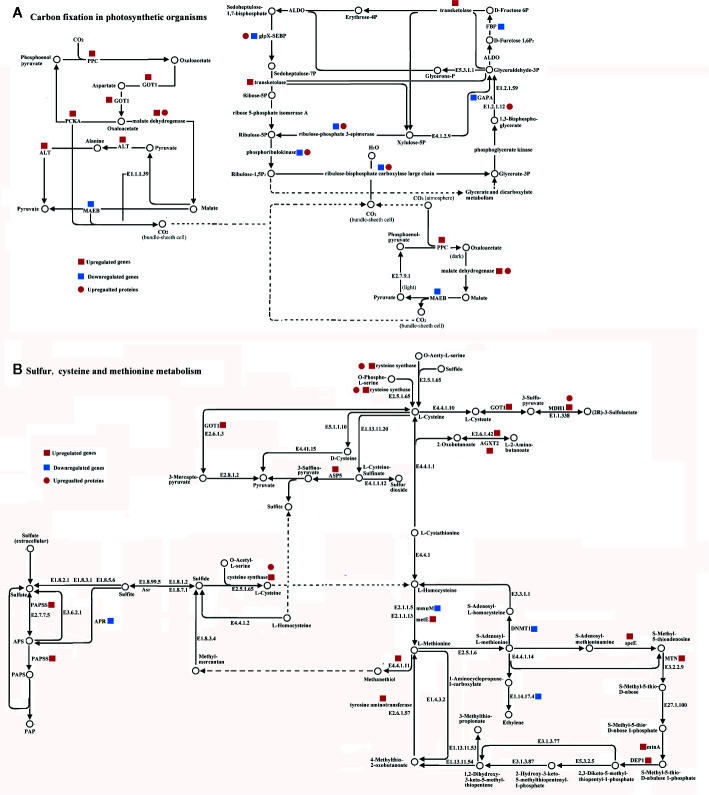
Figure 5.
Integrated pathway of carbon fixation in photosynthetic metabolism pathway (A) and sulfur, cysteine, and methionine metabolism pathway (B) involved in the H2S regulation salt-stress tolerance based on DEGs and DEPs identified in leaves of cucumber. Genes and proteins were mapped on pathway of carbon fixation in photosynthetic or sulfur, cysteine and methionine metabolism using KEGG database, respectively. Red square, blue square, and red circle indicate up-regulated genes, down-regulated genes, and up-regulated proteins, respectively. Abbreviations are as follows: AGXT2, alanine-glyoxylate transaminase; ALT, alanine transaminase; APR, adenylyl-sulfate reductase; ASP5, aspartate aminotransferase; DEP1, methylthioribulose 1-phosphate dehydratase; DNMT1, DNA (cytosine-5)-methyltransferase1; FBP, fructose-1,6-bisphosphatase I; GAPA, glyceraldehyde-3-phosphate dehydrogenase (NADP+); GOT1, glutamic oxaloacetic transaminase; MAEB, malate dehydrogenase (oxaloacetate-decarboxylating)(NADP+); MDH1, malate dehydrogenase; metE, 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase; MTN, 5’-methylthioadenosine nucleosidase; mtnA, methylthioribose-1-phosphate isomerase; PPC, Phosphoenolpyruvate carboxylase; PCKA, phosphoenolpyruvate carboxykinase(ATP); speE, spermidine synthase.