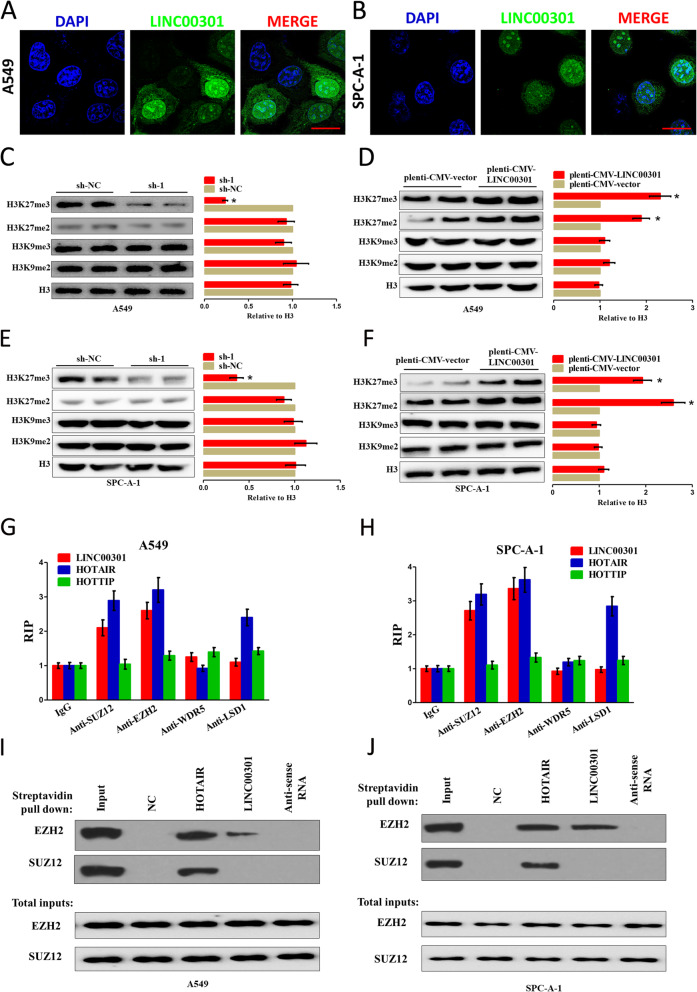
Fig. 6.
LINC00301 positively controls H3K27 trimethylation through interacting with the catalytic subunits for PRC2. a, b RNA FISH assay of LINC00301 in the location at A549 and SPC-A-1 cells. Bar = 30 μm. c–f Representative western blot analysis (left) and quantification of H3 methylation (right), as shown, in A549 and SPC-A-1 cells in LINC00301 KD or OE groups. Methylated H3 level was normalized to total H3 level. Data were shown as mean ± SD. * p < 0.05 by Student’s t-test (versus sh-NC or pLenti-CMV-vector). g, h RIP assay using indicated antibodies, followed by RT–qPCR for LINC00301, HOTAIR, and HOTTIP. Values were normalized to the related IgG RIP groups. Data were shown as mean ± SD. Assays were conducted in triplicate. i, j Representative images of western blot analysis (of n = 2) of a biotinylated RNA streptavidin pull-down assay. Biotinylated full-length (WT) LINC00301 was incubated with A549 or SPC-A-1 cell lysates in RNA-protein binding buffer, followed by streptavidin bead pull-down. PRC2 components such as SUZ12 and EZH2 were exposed by IB in the pull-down products (top set) and total inputs (bottom set). Assays were conducted in triplicate. *p < 0.05, means ± SD was shown. Statistical analysis was performed by Student’s t-test