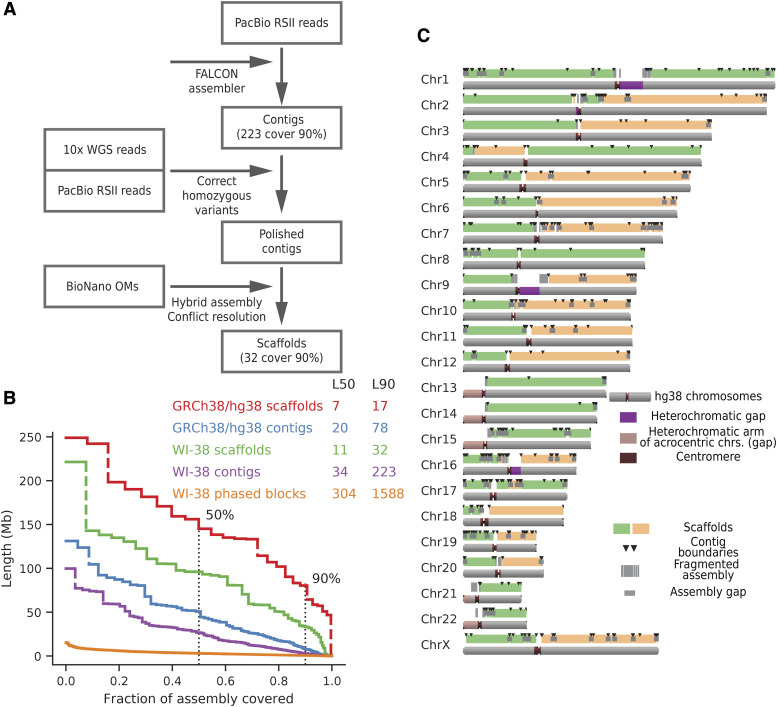
Figure 2.
(A) WI-38 hybrid assembly pipeline. PacBio reads were assembled using FALCON, short errors were corrected using discordant calls from linked reads, and larger assembly errors were corrected using homozygous SV calls from PacBio reads. Contigs were then assembled into scaffolds using optical maps and Bionano Solve pipeline. (B) Relative length distribution of contigs, hybrid scaffolds and phased blocks. Note that while length distribution of contigs and scaffolds was similar to the size distribution in the GRCh38 reference genome, phased blocks were of a significantly shorter length scale (C) Hybrid assembly scaffolds. Coverage of human chromosomes by the scaffolds (green and orange bars mark different neighboring scaffolds) and contigs of the hybrid assembly (black lines denote the breakpoints of contigs) shows that most chromosomes arms were resolved by one major scaffold.