Figure 4.
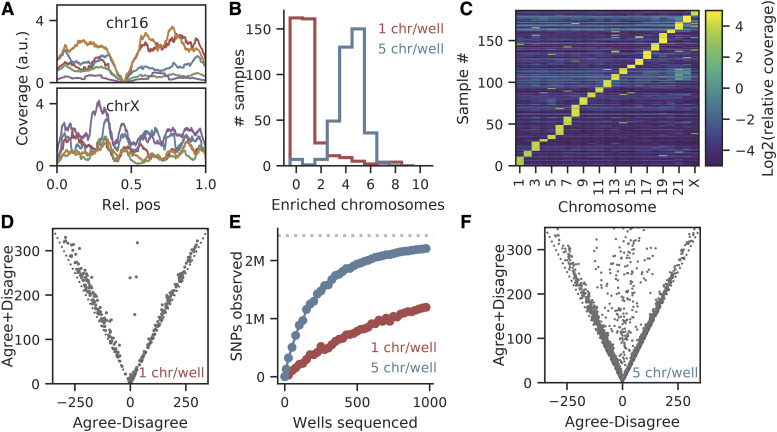
Chromosome sorting isolates individual chromosomes (A) Read coverage. The two panels show the Illumina read coverage from five individual chromosomes mapped across Chr16 (Fig. 4A, upper panel) and ChrX (Fig. 4A, lower panel). Note that the coverage is somewhat uneven on a per Illumina library basis but reasonably evenly distributed in aggregate, except around the centromere for Chr16. (B) Validation of flow-sorting individual chromosomes in 384-well plates. Flow sorting parameters were set so that either one or five separate chromosome events were sorted in wells of a 384-well plate. When mapped to the reference genome, data from plates that targeted one chromosome per well were enriched in reads from one chromosome on average (red line). Wells with 5 chromosome events showed 5 chromosomes on average (blue line). (C) Heatmap of chromosome coverages in each library. Data from single chromosome-sorted libraries were mapped to the reference genome. The X-axis represents the identity of each of the 23 chromosomes while the Y-axis represents the data from each chromosome (data from one chromosome sorted per well in a 384-well plate). As expected, the heatmap shows that reads from each well mapped predominantly to only one chromosome. (D) Pairs of libraries enriched for the same chromosome either agree or disagree on the sampled SNPs. Every point corresponds to a pair of libraries enriched for the same chromosome, Dotted line shows the expected result if SNPs come from the either the same or opposite homologs. (E) Sequencing depth vs. SNP content. As expected, sequencing a greater number of wells leads to a greater number of SNPs in the dataset. This occurred whether one chromosome per well (red dots) or five chromosomes per well (blue dots) were sorted and sequenced. Note significant improvement in SNP detection when pooling five chromosomes per well (F) Data from wells containing five chromosomes still tended to always agree or always disagree on the sampled SNPs. Notation as in (D). But with five chromosomes per well, the number of wells with both haplotypes of a chromosome is higher. If these data are ignored the overall sequencing yield of the number of unique chromosomes per well is still greater than with one chromosome per well.