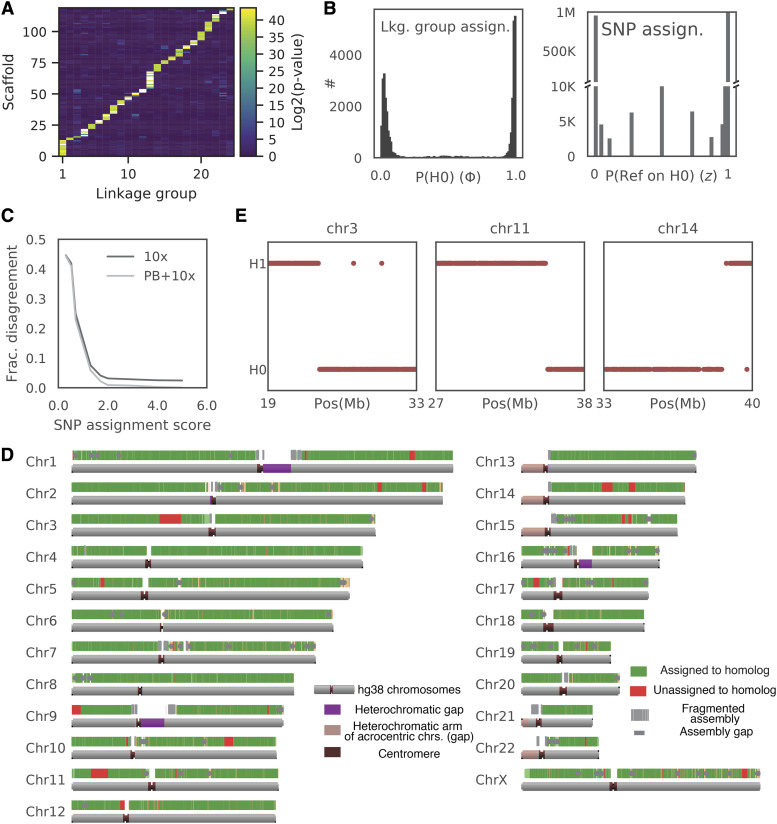
Figure 5.
Phased WI-38 Assembly. Combining SNP phasing from flow-sorted chromosomes with linked-read phased blocks allows accurate assignment of >90% of polymorphisms to their respective homolog. (A) Assignment of scaffolds longer than 100 kb to the linkage group. Heatmap shows probability that a given scaffold belongs to one of the linkage groups. Only scaffolds with a single possible linkage group are shown (see Fig. S7B-C for all scaffolds). (B) Phase of the well (and homolog assignment of the SNP (z) were confidently estimated using EM. Left: Histogram of probabilities that the well is enriched for a certain chromosome contains homolog zero vs. homolog 1 for all (well, enriched chromosome) pairs. Right: Histogram of assignment confidence to a homolog for each SNP. (C) Phase estimates were highly concordant between chromosome-sorted data and the partial phasing. We binned SNPs according to their assignment score -log10(min(z,1-z)) and calculated the fraction of those that disagree with results of partial phasing by linked reads (black line) or of a consensus of linked-reads-derived phased blocks (haplotypes) and FALCON-unzip-derived haplotypes. Note that for over 90% of SNPs with assignment confidence of > 98% there is < 2% discrepancy rate with partial phasing (if the phasing is consistent between 10x and PacBio). (D) Results of assignment of partially phased blocks to homologs. Green - assigned regions, Red - unassigned blocks. Note that most of the unassigned blocks were not assigned due to a phase switch in the middle of the phased block (see(E)). (E) Patterns of agreement. Comparison of the chromosome sorting assignment to the partial phasing in the three largest unassigned phased blocks. Note an apparent breakpoint in homolog assignment that is consistent with a mistake of Longer Ranger pipeline).