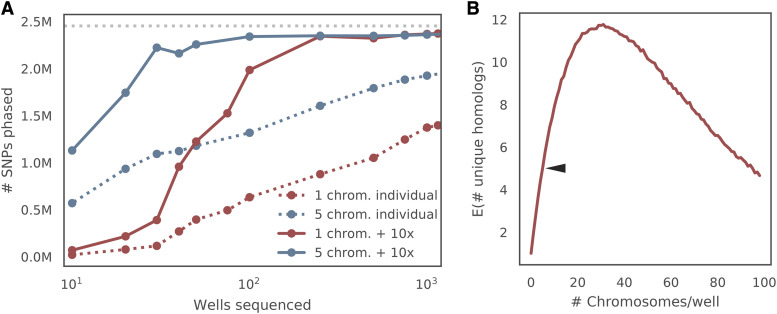
Figure 6.
Optimization of data yield from chromosome sorting (A) Relationship between number of wells sequenced and number of SNPs phased correctly. Using more than one chromosome per well increases the efficiency of sorting and library production. The solid lines show the number of SNPs correctly assigned to homolog if phasing is done using partially phased linked-read blocks, while the dashed lines show the number of SNPs correctly assigned to homolog if phasing is done independently. Red and blue lines show data collected from one chromosome or five chromosomes per well respectively. Note that the correctness of the SNP assignment is determined by using the maximal number of wells sequenced. (B) The number of unique flow-sorted homologs per well. Based on simulations, the optimal data yield (number of unique chromosomes per well) is about 25 chromosomes per well.