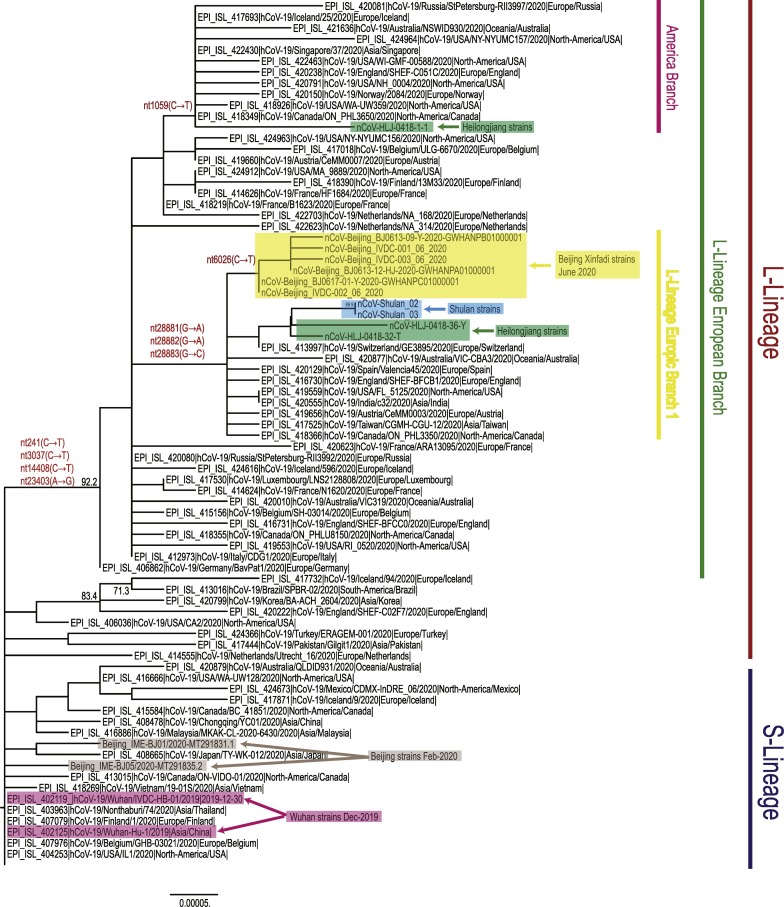
Fig. 1.
Maximum likelihood phylogenetic tree based on the full-length genome sequences of the SARS-CoV-2. The tree was constructed using MEGA (v7.0) software with 1,000 bootstrap replicates. The genomes of the SARS-CoV-2 from Beijing's Xinfadi market are highlighted in shades of yellow. The genomes of the SARS-CoV-2 from Beijing (Feb-2020) and Wuhan (Dec-2019) are highlighted in shades of grey and pink, respectively. The recent reemergence of SARS-CoV-2 in Northeastern China (Shulan and Heilongjiang) that was associated with imported cases are highlighted in shades of blue and green, respectively. S- or L-lineage of the SARS-CoV-2 are marked and colored on the right. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)