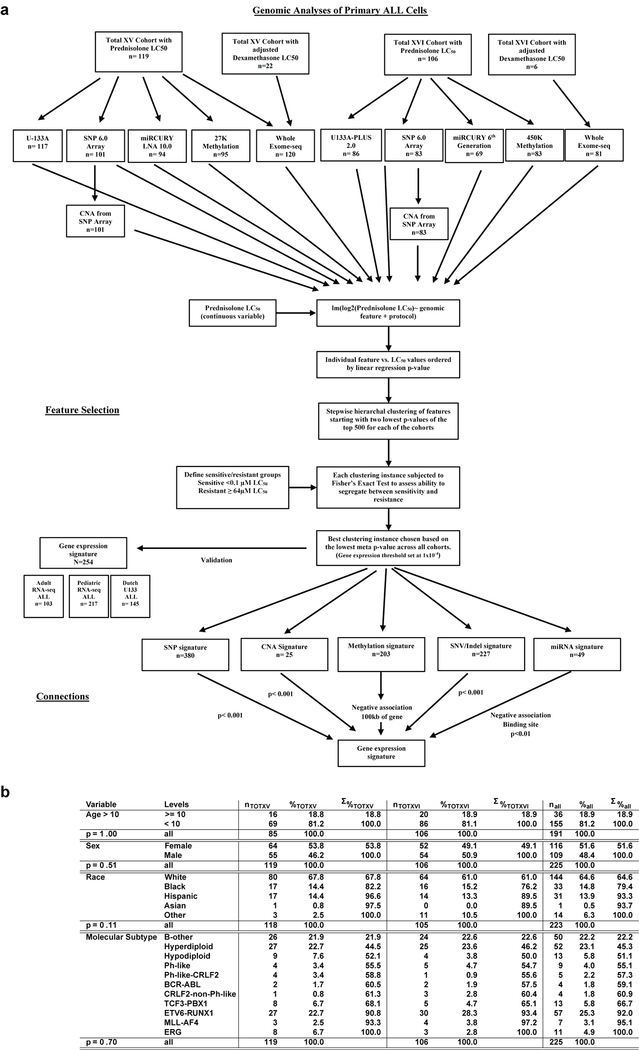
Extended Data Fig. 1. Polygenomic analysis workflow.
(a.) Flowchart depicting cohorts, genomic assays and detailed analysis pipeline for polygenomic analyses of multiple feature types (mRNA, miRNA, DNA methylation, SNVs, CNVs and WES mutations) as determinants of prednisolone sensitivity in patients diagnosed with acute lymphoblastic leukemia (“lm” = linear model). (b.) Table describing age, race, gender and molecular subtype of discovery cohort (n=225 patients) from polygenomic analysis. The P-values represent differences between the discovery cohort enrolled on the two clinical trials (Fisher’s Exact Test p-value; Total 15 and Total 16).