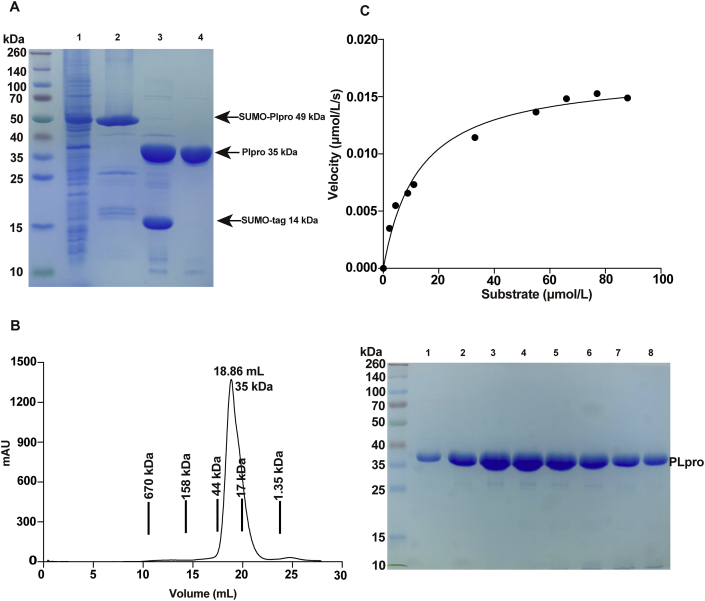
Figure 1.
Production and biochemical characterization of the non-tagged SARS-CoV-2 PLpro. (A) Expression of the non-tagged SARS-CoV PLpro. Lane 1, supernatant of the bacteria lysis overexpressing His-SUMO tagged PLpro; lane 2, eluate from the Ni-NTA resin; lane 3, digestion by Ulp1 overnight; lane 4, flow through from the Ni–NTA column. While the cleaved His-SUMO tag and undigested PLpro species were trapped on the column, non-tagged PLpro proteins were collected. (B) Final purification of PLpro. Left, PLpro was loaded to Superdex 200 10/300GL column, the peak fractions were pooled and concentrated for follow up experiments. Right, SDS-PAGE analysis of the peak fractions. (C) Protease activity of PLpro in cleaving a short synthetic peptide harboring nsp2/nsp3 juncture. The velocity of substrate hydrolysis is plotted as the function of the substrate concentration. The kinetic parameters Km and Vmax were calculated via nonlinear fitting to the Michaelis–Menten equation. The curves were fitted by single measurements, and errors were estimated by curve fitting.