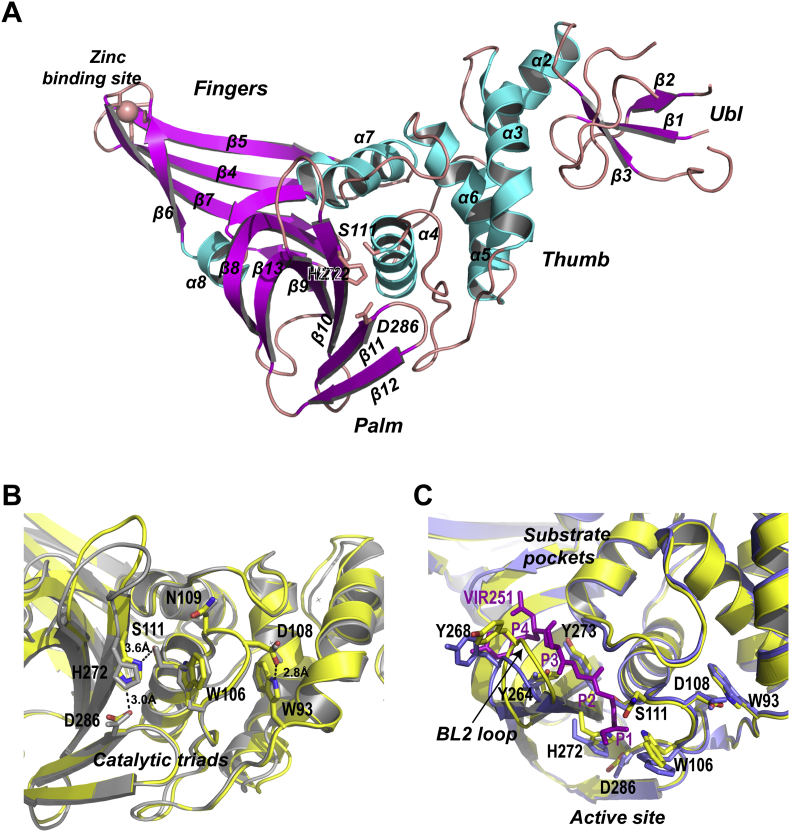
Figure 2.
Crystal structure of the unliganded SARS-CoV-2 PLpro. (A) Ribbon model of the unliganded SARS-CoV-2 PLpro containing C111S mutation. The structure is colored by secondary structural elements, β-strands magenta, α-helices cyan. PLpro subdomains Ub1, Thumb, Finger and Palm are indicated. Four cysteine residues forming the zine finger on the Finger subdomain are shown with stick model. (B) The view of the active site of SARS-CoV-2 PLpro (yellow) superimposed with SARS-CoV PLpro (gray). The catalytic triads C111S-H272-D286, and other catalytically important residues W93 W106, D108 and N109 are shown with stick model. Their counterparts in SARS-CoV PLpro are shown with gray stick model. Key hydrogen bonds are indicated with the dashed lines. (C) Superimposition of the unliganded SARS-CoV-2 PLpro (yellow, PDB ID: 7CJD, C111S mutant) with SARS-CoV-2 PLpro (blue) complexed by peptide inhibitor VIR251 (magenta). The BL2 loop undergoes marked conformational changes upon substrate binding.