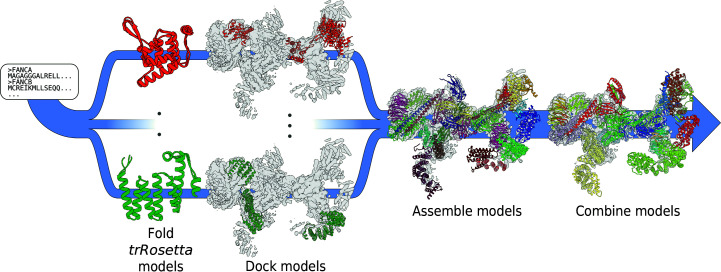
Figure 1.
An overview of the workflow for modeling FAcc. Initially, multiple sequence alignments (MSAs) of all protein sequences are generated, the sequences are segmented into domains using the MSAs and the individual domains are folded using trRosetta. These domains are individually docked into the cryoEM density. Monte Carlo sampling finds the domain assignment that is maximally consistent with the experimental data (electron density, XL-MS etc., Fig. S5). Finally, linkers between domains are sampled and the entire structure is refined.