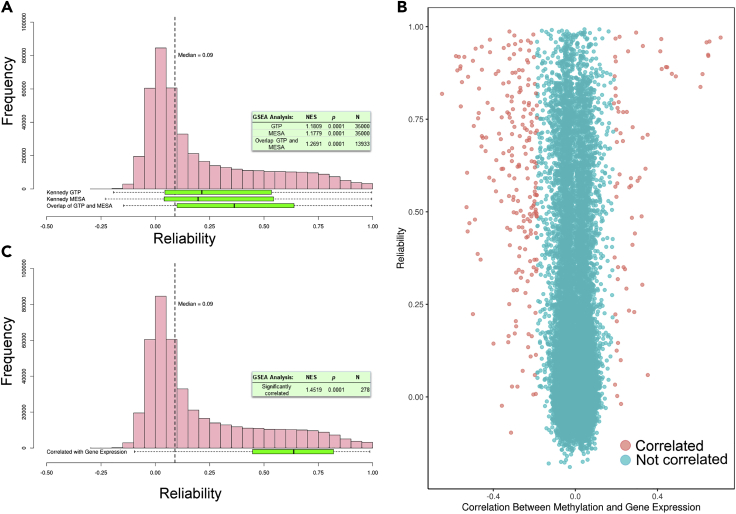
Figure 5.
Reliabilities of Probes Significantly Correlated with Gene Expression Have Higher Reliabilities Than Non-correlated Probes
(A) Distributions of the reliability coefficients of the probes identified as correlated with gene expression by Kennedy et al.40 in the GTP and MESA cohorts (N probes = 36,485 and 114,536, respectively). Probes that are correlated with gene expression in both cohorts are shown in the bottom-most box-and-whisker plot. Boxes correspond to IQR and whiskers extend to 1.5 × IQR. As a reference, the distribution (pink bars) and median (vertical dashed line) of all ∼440,000 probe reliabilities in the E-Risk dataset is shown above the box-and-whisker plots. The text box shows the results of GSEA for the GTP cohort, MESA cohort, and the intersection of both cohorts (NES, normalized enrichment score; N, number of probes). Each cohort's set of significantly correlated DNA methylation probe-gene expression pairs is enriched for reliable probes; pairs that are significantly correlated in both datasets are further enriched.
(B) TSS-localized DNA methylation probe-gene expression probeset correlation (x axis) plotted against DNA methylation probe reliability (y axis) in the Dunedin Study dataset. Probes that were significantly correlated with gene expression are shown in pink (n = 278) and those not correlated are shown in blue.
(C) Distribution of reliabilities of these significantly correlated DNA methylation probes as a box-and-whisker plot. The text box shows the results of GSEA (NES, normalized enrichment score; N, number of probes); DNA methylation probes that were significantly correlated with expression probesets are enriched for reliable probes.