Figure 2.
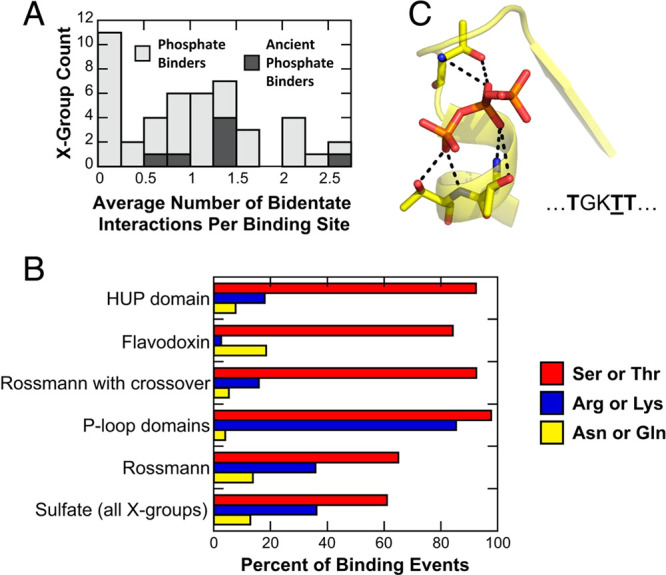
Observed prevalence of bidentate interactions in phosphate binding (where “phosphate” in this case refers broadly to mono-, di-, and triphosphates), based on combined analysis of structural data in the Protein Data Bank59 and evolutionary information in the ECOD database.60,61 X-groups provide the broadest level of classification in ECOD, corresponding to discrete events of evolutionary emergence with no detectable sequence homology or fold identity. Shown here are (A) the frequency of bidentate phosphate binding interactions across all X-groups, including also ancient phosphate binders, and (B) the amino acids involved in forming these bidentate interactions across X-groups and in specific protein folds. Here, it can be seen that Thr and Ser (both prebiotic amino acids) are essential for the formation of bidentate interactions in the N-helix binding mode at the tip N-terminus of an α-helix, an illustrative example of which is shown in the case of the binding of a triphosphate in panel (C). For more details, see ref (63). Reproduced with permission from ref (63). Copyright 2020 National Academy of Sciences.
