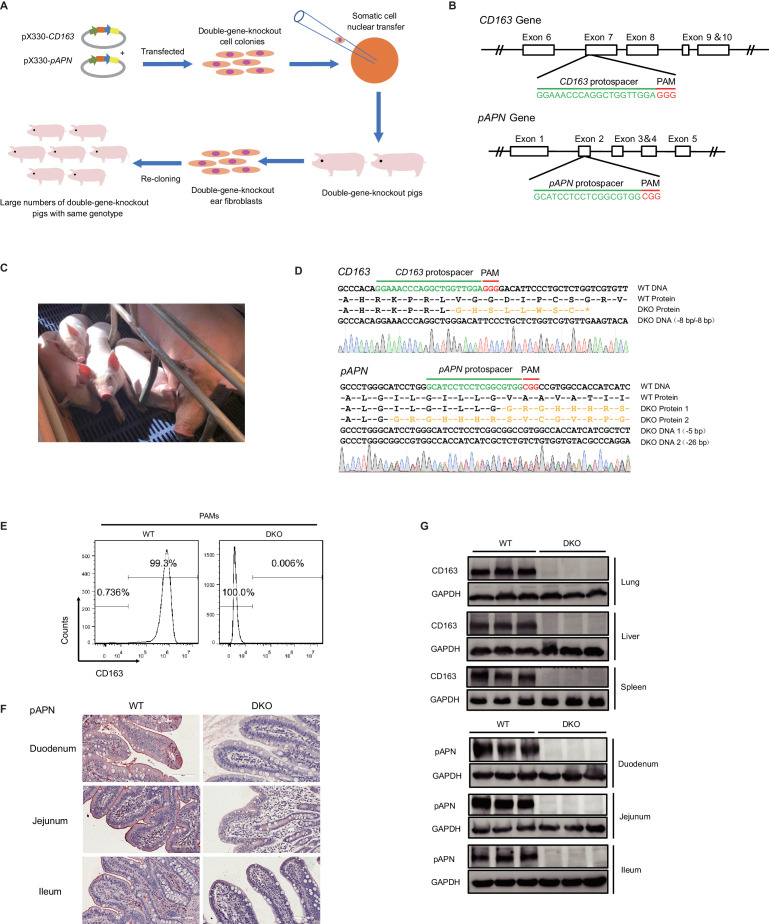
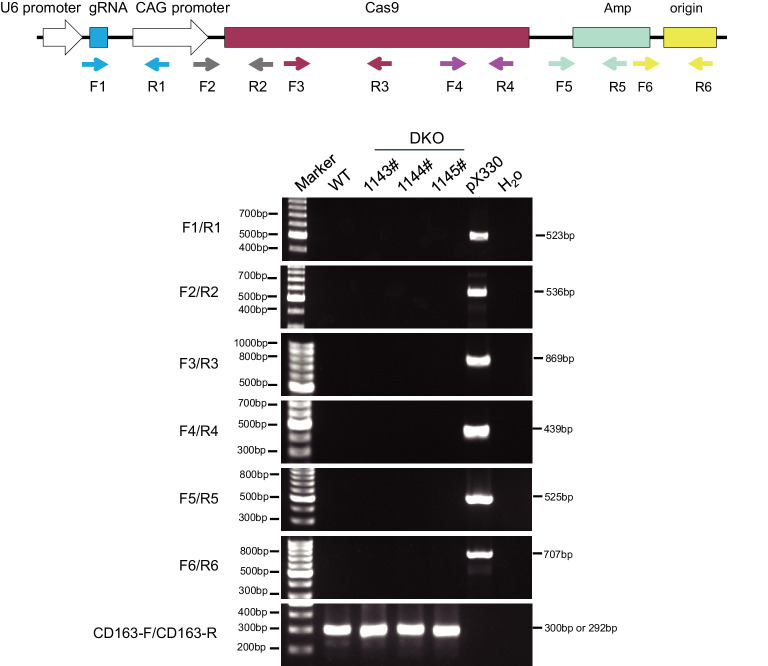
Figure 1. Generation of CD163 and pAPN DKO pigs by CRISPR/Cas9.
(A) Schematic overview of the generation process for DKO cloned pigs. (B) Genetic maps of the CD163 (top) and pAPN (bottom) genes with the location and sequences of the sgRNAs. Exons, white boxes; sgRNA protospacer sequences, green; and PAM sequences, red. (C) Four F0 generation cloned pigs (1085#, 1143#, 1144#, 1145#) aged 1-month-old and a surrogate. (D) Sanger sequencing confirmation of DKO genotypes for three F0 cloned piglets (1143#, 1144#, 1145#). sgRNA protospacer sequences, green; PAM sequences, red; predicted amino acid sequences resulting from frameshift mutations, yellow. (E) Detection of CD163 expression on the surface of PAMs by flow cytometry. (F) Detection of pAPN expression in different small intestine segments by IHC. PAMs and tissues were derived from DKO and WT pigs. (G) Western blot analysis confirmed CD163 and pAPN expression are undetectable in different tissues of DKO pigs (N = 3).