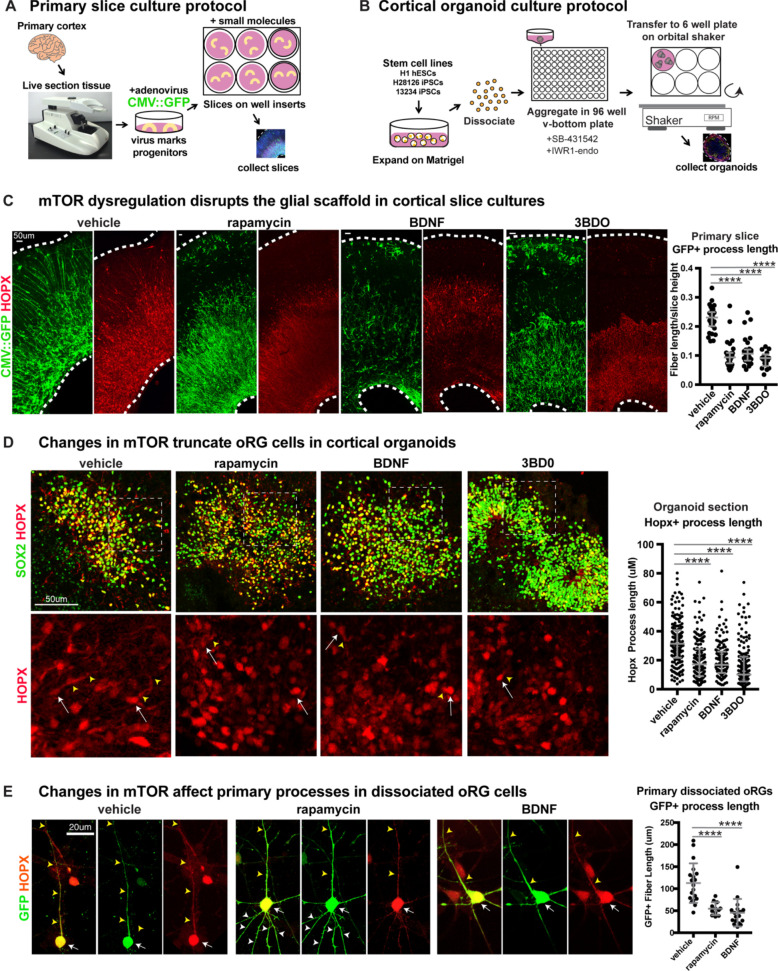
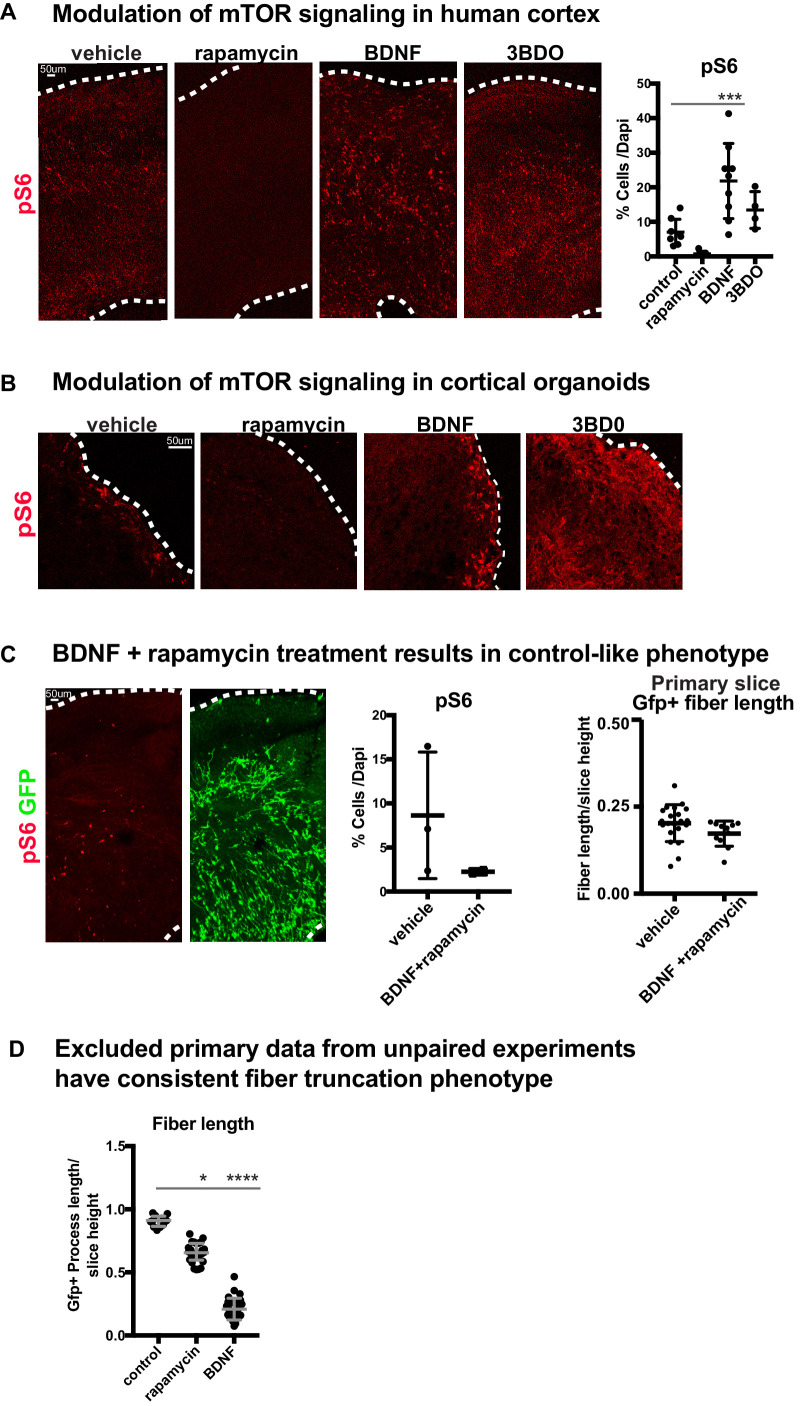
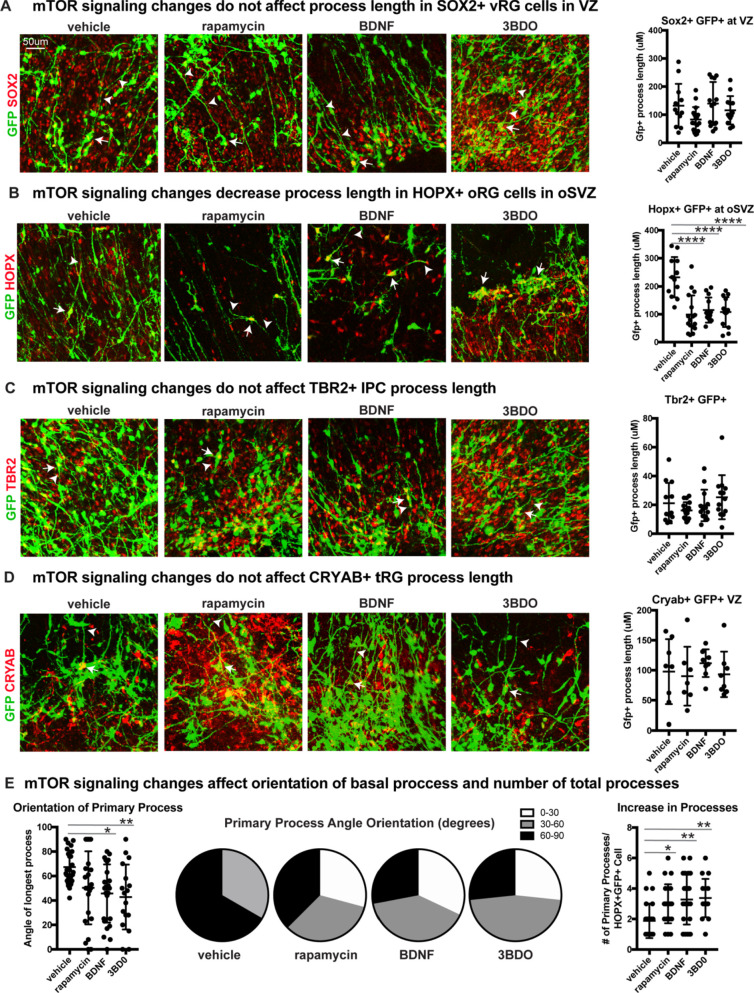
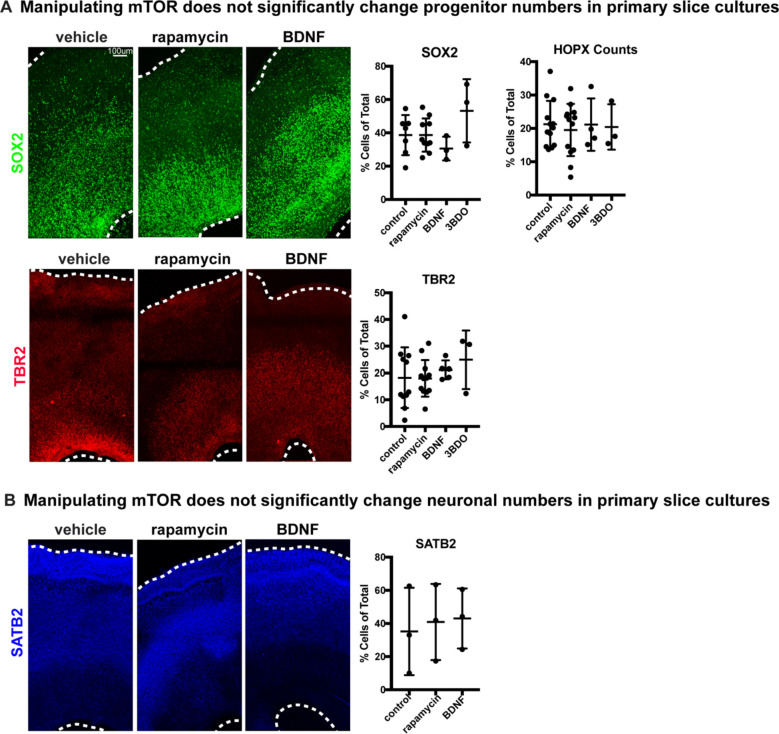
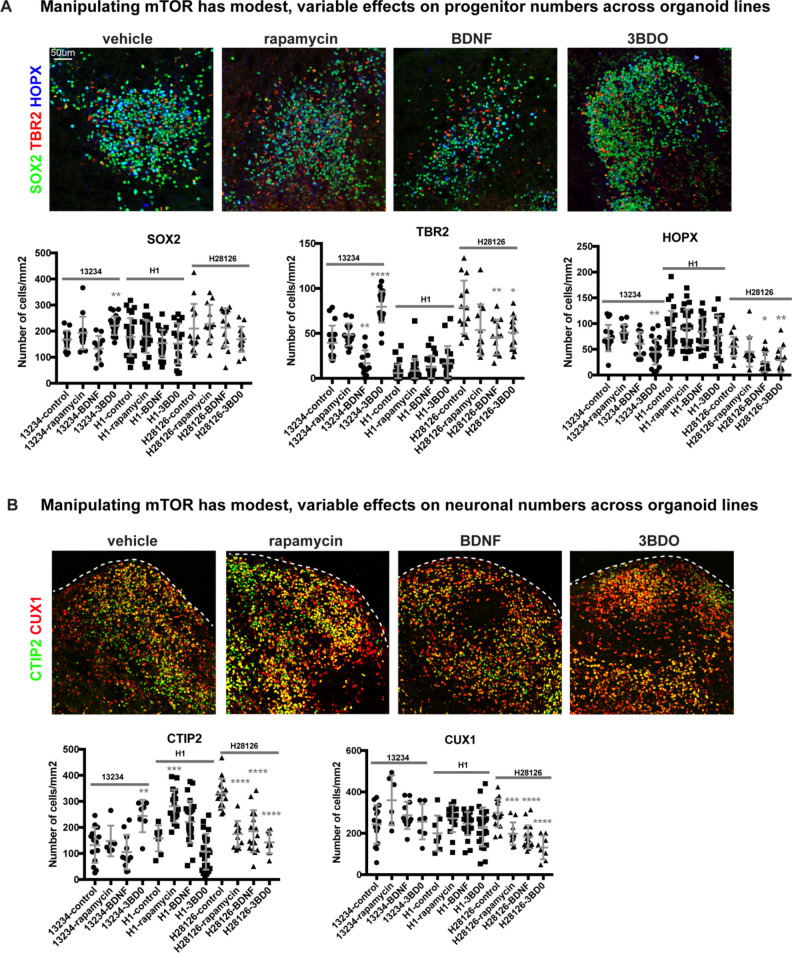
(
A) Three PSC lines were used for organoid studies: 13234, H2826 (iPSCs) and H1 (hESC). The number of cells positive for each marker was counted and then normalized to organoid size. Hyperactivating mTOR signaling has modest effects on progenitor numbers in week 10 organoids, but these effects were not consistent across all iPSC lines or conditions. There was no change to SOX2+ progenitors, except for a small increase in the 3BD0 condition derived from the 13234 iPSC line (SOX2: n = 17 13234 control, n = 26 H1-control, n = 14
H28126-control, n = 14 13234 rapamycin, n = 26 H1 rapamycin, n = 12
H28126 rapamycin, n = 11 13234 BDNF, n = 22 H1 BDNF, n = 14
H28126 BDNF, n = 22 13234 3BD0, n = 18 H1 3BD0, n = 14
H28126 3BD0 sections from >2 organoids/line across three independent experiments; one-way ANOVA with multiple comparisons: 13234 line: control vs rapamycin p=0.386, control vs BDNF p=0.189, control vs 3BD0 **p<0.0027; H1 line: control vs rapamycin p=0.999, control vs BDNF: p=0.630, control vs 3BD0: p=0.627;
H28126 line: control vs rapamycin: p=0.927, control vs BDNF: p=0.999, control vs 3BD0: p=0.417, error bars represent SD). There was an inconsistent change to TBR2+ IPCs in the gain of function conditions in two iPSC lines, where both increase, decrease, and no change in numbers were observed (TBR2: n = 17 13234 control, n = 26 H1-control, n = 14
H28126-control, n = 14 13234 rapamycin, n = 26 H1 rapamycin, n = 12
H28126 rapamycin, n = 11 13234 BDNF, n = 22 H1 BDNF, n = 14
H28126 BDNF, n = 22 13234 3BD0, n = 18 H1 3BD0, n = 14
H28126 3BD0 sections; one-way ANOVA with multiple comparisons: 13234 line: control vs rapamycin p=0.433, control vs BDNF **p>0.004, control vs 3BD0 ***p<0.0001; H1 line: control vs rapamycin p=0.913, control vs BDNF p=0.404, control vs 3BD0 p=0.070;
H28126 line: control vs rapamycin p=0.05, control vs BDNF **p<0.004, control vs 3BD0 *p<0.018, error bars represent SD). Overall, there was little effect on HOPX+ oRG number, but a small decrease was observed in mTOR activation conditions in two lines (n = 17 13234 control, n = 26 H1-control, n = 14
H28126-control, n = 14 13234 rapamycin, n = 26 H1 rapamycin, n = 12
H28126 rapamycin, n = 11 13234 BDNF, n = 22 H1 BDNF, n = 14
H28126 BDNF, n = 22 13234 3BD0, n = 18 H1 3BD0, n = 14
H28126 3BD0 sections from >2 organoids/line across three independent experiments; one-way ANOVA with multiple comparisons: 13234 line: control vs rapamycin: p=0.599, control vs BDNF p=0.579, control vs 3BD0 **p<0.004; H1 line: control vs rapamycin p=0.9950, control vs BDNF p=0.956, control vs 3BD0 p=0.726;
H28126 line: control vs rapamycin p=0.717, control vs BDNF **p<0.007, control vs 3BD0 *p<0.022, error bars represent SD). Organoid images shown are derived from the 13234 iPSC line. (
B) There are modest and inconsistent changes to neuronal populations in organoids. There is a small increase in CTIP2+ cells in the rapamycin treatment group of the H1 stem cell line and the 3BD0 group in the 13234 line, while there is a decrease in the
H28126 line in all mTOR manipulation groups. (CTIP2: n = 15 13234 control, n = 8 H1-control, n = 16
H28126-control, n = 6 13234 rapamycin, n = 17 H1 rapamycin, n = 11
H28126 rapamycin, n = 11 13234 BDNF, n = 25 H1 BDNF, n = 17
H28126 BDNF, n = 7 13234 3BD0, n = 24 H1 3BD0, n = 8
H28126 3BD0 sections from >2 organoids/line across three independent experiments; one-way ANOVA with multiple comparisons: 13234 line: control vs rapamycin p=0.959, control vs BDNF p=0.733, control vs 3BD0 **p<0.004; H1 line: control vs rapamycin ***p<0.0006, control vs BDNF p=0.130, control vs 3BD0 p=0.291,
H28126 line: control vs rapamycin, BDNF, 3BD0 ****p<0.0001, error bars represent SD). There is a decrease in CUX1+ neurons across mTOR manipulations in the
H28126 line, but no change in organoid derived from other lines (CUX1: n = 15 13234 control, n = 8 H1-control, n = 16
H28126-control, n = 6 13234 rapamycin, n = 17 H1 rapamycin, n = 11
H28126 rapamycin, n = 11 13234 BDNF, n = 25 H1 BDNF, n = 17
H28126 BDNF, n = 7 13234 3BD0, n = 24 H1 3BD0, n = 8
H28126 3BD0 sections from >2 organoids/line across three independent experiments; one-way ANOVA with multiple comparisons: 13234 line: control vs rapamycin p=0.052, control vs BDNF p=0.617, control vs 3BD0 p=0.988; H1 line: control vs rapamycin p=0.130, control vs BDNF p=0.439, control vs 3BD0 p=0.763;
H28126 line: control vs rapamycin: ***p<0.0005, control vs BDNF, 3BD0 ****p<0.0001, error bars represent SD). Organoid images shown are derived from the H1 ESC line.