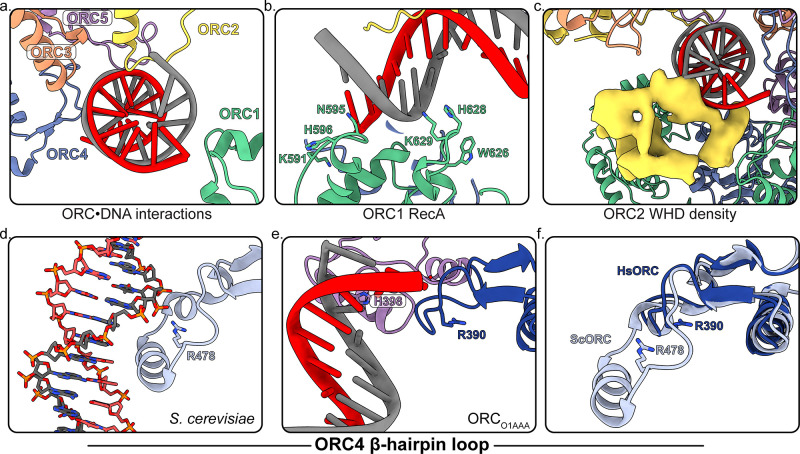
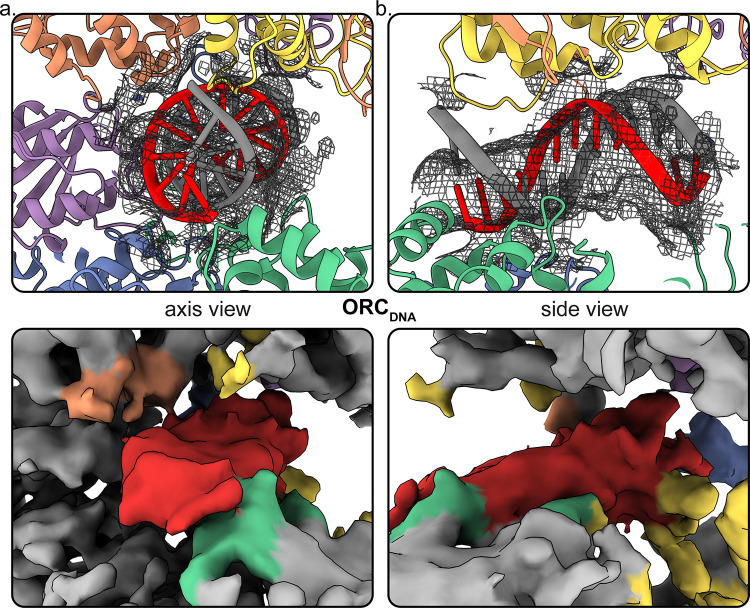
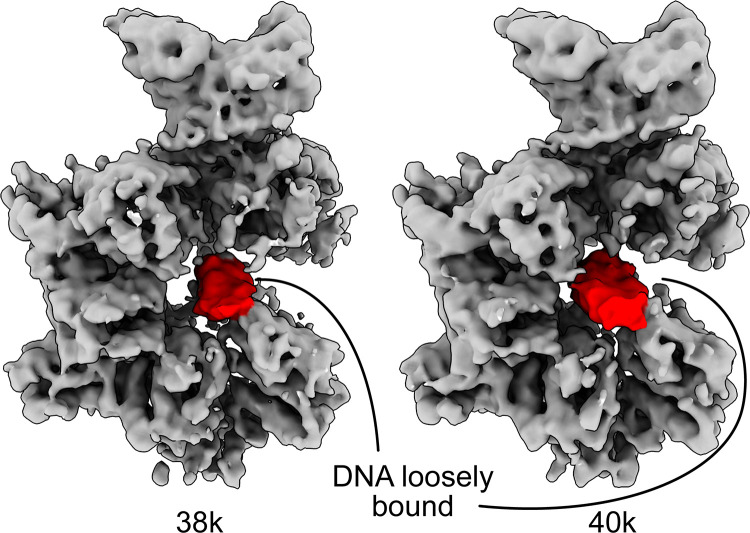
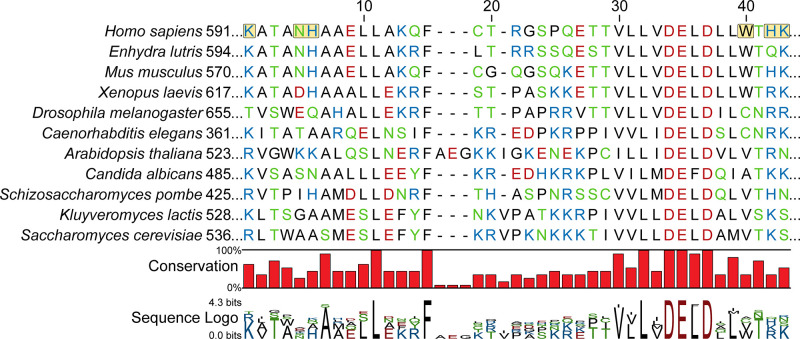
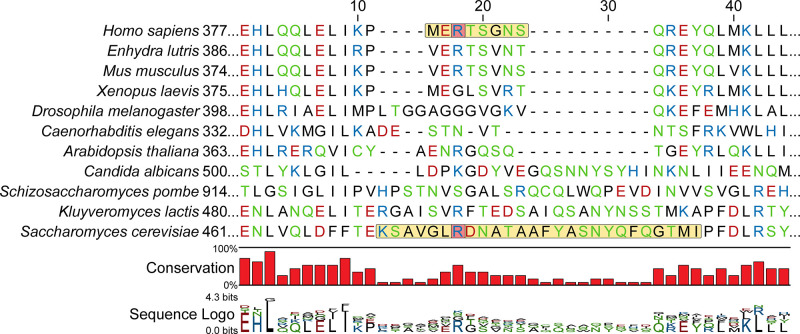
Figure 4. Several regions of ORC contact the endogenous DNA in ORCDNA.
(a) ORC subunits 1–5 contain regions with positively-charged residues proximal to the DNA. (b) The ORC1 RecA-fold contains several positively-charged residues in close proximity to DNA. (c) The ORC2 WHD, shown as a density map due to the lower resolution of this domain, moves to interact with the DNA in the core of the complex. (c) The β-hairpin loops of ORC4 and ORC5 contain positively-charged residues proximal to the DNA. The DNA depicted consists of non-specific sequence. (d) The ORC4 β-hairpin loop of S. cerevisiae contains an α-helix that inserts into the major groove of DNA making sequence-specific interactions with DNA bases. (e) The β-hairpin loop in ORCDNA does not contain an α-helix. However, ORC4 R390 in humans may interact with the DNA backbone similar to ORC4 R478 in S. cerevisiae. (f) The superimposition of the ScORC and HsORC β-hairpin loops illustrates the significant difference between the regions.