Abstract
The sensing modules for analyzing miRNAs or the endonucleases consist of tetrahedra functionalized with three different fluorophore–quencher pairs in spatially quenched configurations and hairpin units acting as recognition elements for the analytes. Three different miRNAs (miRNA-21, miRNA-221, and miRNA-155) or three different endonucleases (Nt.BbvCI, EcoRI, and HindIII) uncage the respective hairpins, leading to the switched-on fluorescence of the respective fluorophores and to the multiplex detection of the respective analytes. In addition, a tetrahedron module for the multiplexed analysis of aptamer ligand complexes (ligands = ATP, thrombin, VEGF) is introduced. The module includes edges modified with three spatially separated fluorophore–quencher pairs that were stretched by the respective aptamer strands to yield a switched-on fluorescent state. Formation of the respective aptamer ligands reconfigures the edges into fluorophore-quenched caged-hairpin structures that enable the multiplexed analysis of the aptamer–ligand complexes. The facile permeation of the tetrahedra structures into cells is used for the imaging of MCF-7 and HepG2 cancer cells and their discrimination from normal epithelial MCF-10A breast cells.
Keywords: fluorescence, sensors, VEGF, thrombin, DNA nanotechnology
The development of parallel and multiplexed sensing platforms for the detection of microRNAs, aptamer–ligands, nuclease activities, or DNA has utmost significance for the detection and progress of diseases,1−3 gene profiling,4 environmental monitoring,5,6 detection of pathogens,7 and biowarfare agents.8 Different analytical schemes for the multiplexed electrochemical and optical analysis of targets were reported. For example, bar-coded metal microrods generated in alumina channels were suggested for high-throughput analysis.9 Graphene oxide10 or transition metal dichalcogenide nanosheets11 were applied for the multiplexed fluorescence analysis of genes or aptamer–ligands. Also, the size-controlled luminescence properties of semiconductor quantum dots12,13 or metal nanoclusters, such as Ag nanoclusters,14,15 were used to develop multiplexed detection schemes. Hybrid conjugates consisting of hemin/G-quadruplex and different-sized semiconductor quantum dots were applied for the multiplexed analysis of genes16 and aptamer–ligand complexes17 using chemiluminescence resonance energy transfer (CRET) as the readout signal. Multiplexed electrochemical detection of analytes was achieved by the application of metal nanoparticles18 or semiconductor nanoparticles13,19,20 as labeling codes for the sensing events. Also, microfluidic electrochemical aptamer-based assays were integrated on chips for the amplified and multiplexed analysis of aptamer–ligand complexes.21 In addition, DNA machineries such as the hybridization chain reaction22 or rolling circle amplification process,23,24 or the use of multigene DNA scaffolds and polymerization/nicking enzyme machineries, for the autonomous synthesis of DNAzyme-generating specific fluorescent readout signals were used for multiplexed sensing.25
The rapidly developing area of DNA nanotechnology26,27 introduced different nucleic-acid-functionalized nano- and microhybrid carriers, such as nanoparticles,28−31 metal–organic framework particles,32−35 and microcapsules,36,37 acting as sense-and-treat systems. Also, the dictated hybridization of complementary nucleic acids led to the assembly of supramolecular nanostructures such as origami systems38,39 or oligomerized crossover junction units.40,41 The DNA tetrahedron represents a unique three-dimensional nanostructure.42−44 The possibility to modify the corners of the tetrahedron with different chemical functions, such as different fluorophores, and the feasibility to functionalize the edges of the tetrahedron with nucleic acid functionalities, such as stimuli-responsive or DNAzyme units, allow the programmed reconfiguration of the tetrahedron edges.45 Indeed, these unique properties of DNA tetrahedra nanostructures were used for the development of electrochemical DNA sensors.46 In addition, DNA tetrahedra nanostructures revealed effective permeation into living cells,47,48 and these properties were applied to sense intracellular ingredients,49 to image cells,50 and to use the tetrahedron scaffold as a functional unit to amplify miRNA detection in cells.51
In the present study, we introduce the DNA tetrahedron module as a versatile unit for diverse parallel and multiplexed sensing applications. We describe the engineering of the edges of the tetrahedra with three different fluorophore–quencher pairs and with predesigned hairpin structures and present the use of the tetrahedra as functional modules for the parallel and multiplexed sensing of different analytes. Specifically, we apply the modules for the multiplexed sensing of miRNAs, of enzymes cleaving DNA (endonucleases), and of ligand–aptamer complexes. In addition, the facile permeation of the tetrahedra structures into cells is used for the imaging of MCF-7 and HepG2 cancer cells and their distinction from normal epithelial MCF-10A breast cells by the fluorophore/quencher-functionalized sensing module.
Results and Discussion
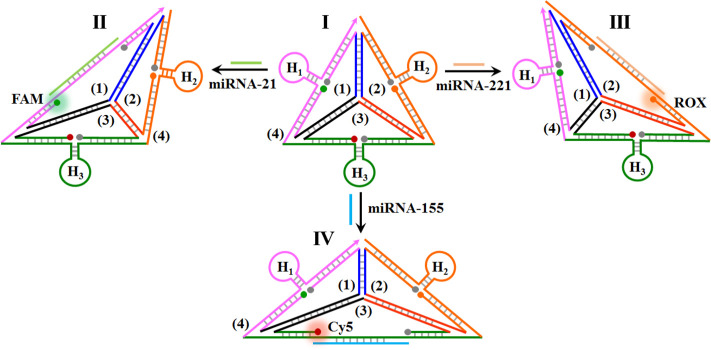
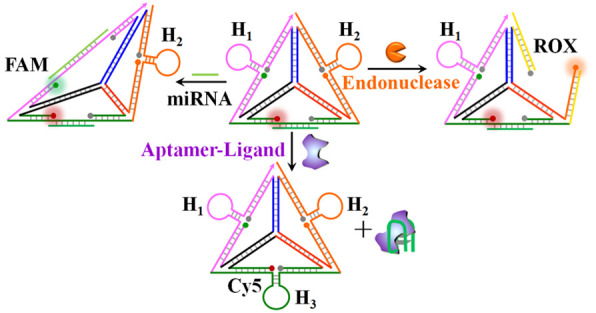
MicroRNAs (miRNAs) are short noncoding RNA molecules (21–24 bases) that post-transcriptionally regulate gene expression in various cellular processes.52,53 Particularly misregulated expression of miRNAs is closely correlated with cancer development and progression, thus highlighting the importance of detection of miRNAs for clinical diagnostics.54−56Scheme 1 outlines the assembly of the DNA tetrahedron structure for the multiplexed analysis of three different miRNAs. Three nucleic acid strands, 1, 2, and 3, presented schematically as three quasi-triangle structures, are interhybridized by appropriate base-paired domains, and the resulting supramolecular 1/2/3 structure is further rigidified by a peripheral strand 4 that hybridizes with the single-strand domains of the quasi-triangle edges of 1, 2, and 3. The interhybridized composite 1/2/3/4 forms the resulting tetrahedron nanostructure, state I. The single-strand quasi-triangle edges of 1, 2 and 3 are modified at their 3′- and 5′-ends with fluorophore/quencher pairs (for 1, FAM/BHQ1; for 2, ROX/BHQ2; for 3, Cy5/BHQ2). The peripheral rigidifying strand, stabilizing the quasi-triangle-shaped units, includes three hairpin components, H1, H2, and H3, where the stem regions of the hairpins are engineered at spatial positions in proximity to the fluorophore/quencher units associated with the quasi-triangles 1, 2, and 3, respectively. Under these conditions, the fluorophores associated with edges of the DNA tetrahedron are quenched. The hairpins H1, H2, and H3 include, in the loop regions, the recognition sequences for miRNA-21, miRNA-221, and miRNA-155, respectively. The method to use the hairpin-functionalized tetrahedron as a functional scaffold for sensing the three different miRNAs, and particularly for the multiplexed analysis of the miRNAs, is also addressed in Scheme 1. Treatment of the rigidified tetrahedron scaffold, state I, with miRNA-21 results in the opening of hairpin H1 and the distortion of the scaffold, thus forming state II. The formation of the duplex between H1 and miRNA-21 distorts the shape of the H1-containing edge, resulting in the spatial separation of the fluorophore/quencher pair (FAM/BHQ1) and the enhanced fluorescence of the FAM (λ = 516 nm). Note that miRNA-21 does not interact with hairpins H2 and H3, and thus, the fluorescence of the fluorophores in proximity to hairpins H2 and H3 is not affected. Treatment of the sensing platform with miRNA-221 opens hairpin H2 and generates state III, where the fluorophore/quencher pair (ROX/BHQ2) is separated, and the fluorescence of ROX, λ = 600 nm, is intensified. Also, subjecting the sensing platform to miRNA-155 leads to the opening of hairpin H3 and to the formation of state IV, where the tetrahedron edge consisting of the fluorophore/quencher pair (Cy5/BHQ2) is stretched. This results in the intensified fluorescence of Cy5, λ = 663 nm. Note that the sensing scaffold, state I, provides a functional nanostructure for the multiplexed analysis of the miRNAs. In the presence of any combination of two miRNAs, or in the presence of all three miRNAs, the respective hairpins are anticipated to be unlocked, leading to the intensified fluorescence of the fluorophores associated with the respective hairpin-modified edges.
Scheme 1. Schematic Multiplexed Analysis of Three Different miRNAs by a Functional DNA Tetrahedra Module.
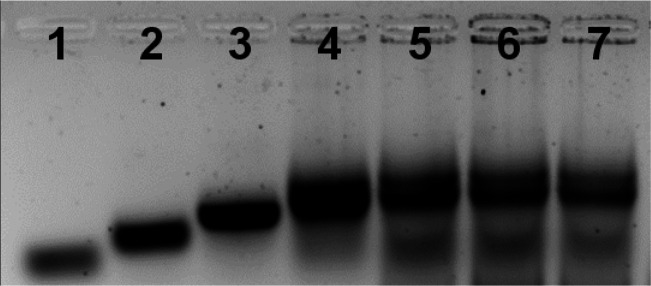
In the first step, the formation of the intact tetrahedron scaffold, state I, and the association of miRNA-155, miRNA-21, and miRNA-221 were confirmed by electrophoretic separation (Figure 1). In lane 1, the band of the quasi-triangle strand 1 is presented. Lanes 2, 3, and 4 show the stepwise construction of the interhybridized structures 1/2, 1/2/3, and 1/2/3/4. The stepwise increase in the molecular weight of the structures leads to slower migration rates. Lanes 5, 6, and 7 depict the bands corresponding to the resulting miRNAs hybridized with the tetrahedron sensing scaffold. It should be noted that the gel electrophoresis shown in Figure 1 demonstrates the intact structure of the tetrahedra modified with each of the miRNA. The electrophoretic studies were further complemented with agarose gel electrophoretic separation, examining the multiplexed detection of the miRNAs (Figure S1 and accompanying discussion).
Figure 1.
Agarose gel electrophoresis (2%) analysis of the construction of the tetrahedron sensing modules described in Scheme 1. Lane 1, strand 1; lane 2, duplex strand 1 + 2; lane 3, Y-shaped structure consisting of strands 1 + 2 + 3; lane 4, intergrated tetrahedra module composed of 1 + 2 + 3 + 4; lane 5, intergrated tetrahedra modified with miRNA-21; lane 6, intergrated tetrahedra carrying miRNA-155; lane 7, intergrated tetrahedra carrying miRNA-221.
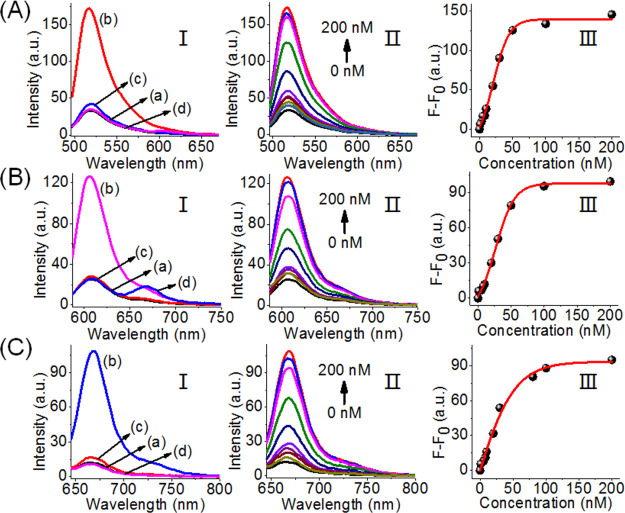
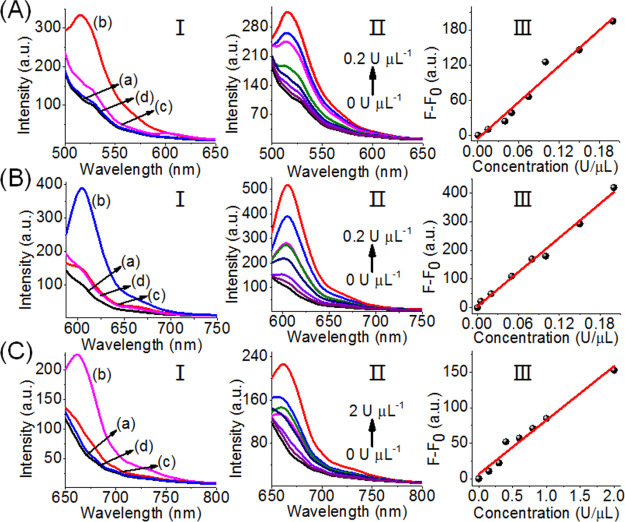
Figure 2 shows the selective optical (fluorescence) sensing of the three miRNAs by the sensing scaffold, state I. Figure 2A, panel I, shows the fluorescence intensities of FAM before the addition of miRNA-21, curve (a), and after the addition of miRNA-21, 0.2 μM, curve (b). For comparison, curves (c) and (d) show the fluorescence intensities of the sensing platform upon addition of the miRNA-155 and miRNA-221. The fluorescence of FAM is not affected by miRNA-155 and miRNA-221, and the selective intensified fluorescence of FAM is observed upon the miRNA-21-driven unlocking of hairpin H1. Figure 2A, panel II, shows the fluorescence spectra of FAM upon the treatment of the sensing platform, with different concentrations of miRNA-21. As the concentration of miRNA-21 increases, the fluorescence intensity increases consistently with the enhanced opening of hairpin H1 associated with the sensing scaffold. Figure 2A, panel III, depicts the calibration curve corresponding to the fluorescence intensities of FAM in the presence of variable concentrations of miRNA-21. The fluorescence intensity of FAM associated with the scaffold levels off at a miRNA-21 concentration corresponding to ∼50 nM. The detection limit for sensing miRNA-21 corresponds to 1 nM. Similarly, Figure 2B, panel I, shows the fluorescence spectra of ROX associated with the sensing scaffold in the absence of miRNA-221, curve (a), and in the presence of miRNA-221, 0.2 μM, curve (b). Subjecting the sensing platform to miRNA-221 results in the opening of H2 and the formation of the asymmetric tetrahedron, state III, where the fluorophore ROX is spatially separated from the quencher unit. The spatial separation of the fluorophore from the quencher leads to the enhanced fluorescence of ROX. Figure 2B, panel I, shows the fluorescence of the fluorophore ROX upon addition of miRNA-21, curve (c), and miRNA-155, curve (d). Only the background fluorescence of the sensing scaffold is observed, implying that the foreign miRNA-21 and miRNA-155 do not interfere or perturb the sensing of miRNA-221. Figure 2C, panel I, depicts the fluorescence spectra of Cy5 associated with the tetrahedron sensing platform in the absence of miRNA-155, curve (a), and in the presence of miRNA-155, 0.2 μM, curve (b). The resulting fluorescence of Cy5 generated by the scaffold upon addition of miRNA-155 is consistent with the opening of hairpin H3 and the spatial separation of the fluorophore/quencher pair, state IV. As before, subjecting the tetrahedron sensing platform to the foreign miRNA-21 and miRNA-221 does not affect the florescence of Cy5, and only the background fluorescence of Cy5 is observed, curves (c) and (d), respectively. The fluorescence spectra of Cy5 of the sensing platform, in the presence of variable concentrations of miRNA-155, are shown in Figure 2C, panel II, and the calibration curve corresponding to the fluorescence intensity of Cy5, in the presence of variable concentrations of miRNA-155, is presented in Figure 2C, panel III. As the concentrations of miRNA-155 increase, the fluorescence of Cy5 is intensified, and it levels off to a saturation level at a miRNA-155 concentration of ∼100 nM. The detection limit for sensing miRNA-155 corresponds to 1 nM. The results indicate that the tetrahedron sensing module, state I, allows the parallel selective detection of the three miRNAs.
Figure 2.
Analysis of three miRNAs by the DNA tetrahedron module described in Scheme 1. (A) Analysis of miRNA-21. Panel I: fluorescence spectra of FAM (a) in the absence of added miRNA-21, (b) upon the addition of miRNA-21, 0.2 μM, (c) upon addition of miRNA-221, 0.2 μM, (d) upon addition of miRNA-155, 0.2 μM. Panel II: fluorescence changes of FAM upon the addition of variable concentrations of miRNA-21. Panel III: derived calibration curve corresponding to the fluorescence changes of FAM at different concentrations of miRNA-21. (B) Analysis of miRNA-221. Panel I: fluorescence spectra of ROX (a) in the absence of added miRNA-221, (b) upon the addition of miRNA-221, 0.2 μM, (c) upon addition of miRNA-21, 0.2 μM, (d) upon addition of miRNA-155, 0.2 μM. Panel II: fluorescence changes of ROX upon the addition of variable concentrations of miRNA-221. Panel III: derived calibration curve corresponding to the fluorescence changes of ROX at different concentrations of miRNA-221. (C) Analysis of miRNA-155. Panel I: fluorescence spectra of Cy5 (a) in the absence of added miRNA-155, (b) upon the addition of miRNA-155, 0.2 μM, (c) upon addition of miRNA-21, 0.2 μM, (d) upon addition of miRNA-221, 0.2 μM. Panel II: fluorescence changes of Cy5 upon the addition of variable concentrations of miRNA-155. Panel III: derived calibration curve corresponding to the fluorescence changes of Cy5 at different concentrations of miRNA-155.
The selective fluorescence of the three fluorophores associated with the tetrahedron scaffold suggests that the sensing module could be applied for the multiplexed analysis of the three miRNAs. Figure 3 demonstrates the multiplexed analysis of the set of miRNAs. The fluorescence changes observed upon subjecting the sensing module to miRNA-21 and miRNA-155 are presented in panel I. The fluorescence intensities of FAM (reporter for miRNA-21) and Cy5 (reporter for miRNA-155) are intensified, whereas no fluorescence change of ROX (reporter for miRNA-221) is observed. Multiplexed analysis of miRNA-155 and miRNA-221 is reflected by high fluorescence changes of Cy5 and ROX and lack of any fluorescence changes of FAM. In addition, panel III demonstrates the multiplexed analysis of miRNA-221 and miRNA-21. Finally, panel IV depicts the multiplexed analysis of all three miRNAs. In this experiment, the sensing module is subjected to miRNA-155, miRNA-21, and miRNA-221. The fluorescence intensities of FAM, ROX, and Cy5 are intensified, demonstrating that the parallel multiplexed detection of all three miRNAs is, indeed, feasible.
Figure 3.
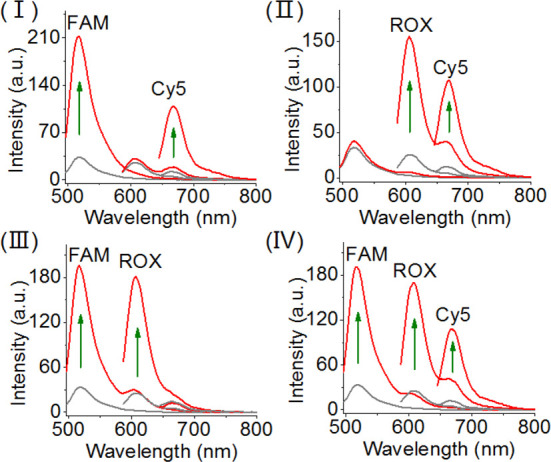
Multiplexed analysis of the three miRNA targets, miRNA-21, miRNA-221, and miRNA-155 by the DNA tetrahedron sensing module. Panel I: sensing of miRNA-21 and miRNA-155, switches on the fluorescence of FAM and Cy5. Panel II: sensing miRNA-221 and miRNA-155 by the sensing module, switches on the fluorescence of ROX and Cy5. Panel III: sensing miRNA-21 and miRNA-221 by the sensing module, switches on the fluorescence of FAM and ROX. Panel IV: multiplexed sensing of all three miRNAs (miRNA-21, miRNA-221, and miRNA-155), switches on the fluorescence of FAM, ROX, and Cy5. The concentrations of each of the miRNAs in the systems correspond to 0.2 μM.
Although the DNA tetrahedra structure provides an elegant nanostructure for the multiplexed analysis of miRNAs, one could argue that the same analytical challenge may be demonstrated by other mixtures of sensing probes, such as a mixture of three different hairpins modified with three different fluorophore/quencher pairs. We feel, however, that the applications of an integrated functional tetrahedron nanostructure for the multiplexed parallel analysis of three miRNAs could reveal significant biosensing and imaging advantages due to the easy, high-yield, permeation of the DNA tetrahedra into cells. This is demonstrated in Figure 4 with the multiplexed analysis of miRNA-21 and miRNA-155 in two types of cancer cells (e.g., MCF-7 breast cancer cells, HepG2 liver cancer cells), in comparison to normal epithelial breast cells MCF-10A. The MCF-7 cells include the miRNA-21 and miRNA-155 as biomarkers,57 and the HepG2 cells overexpress miRNA-21.58 Accordingly, the different cells were treated with the tetrahedra composite shown in Scheme 1 that includes three quenched fluorophores, FAM, Cy5, and ROX. Figure 4, entry I, shows the bright-field image and confocal fluorescence microscopy images of the MCF-7 cell recorded at the excitation wavelengths of the different fluorophores associated with the tetrahedra. The green FAM fluorescence corresponding to miRNA-21 and red Cy5 fluorescence corresponding to miRNA-155 are observed, and the ROX fluorescence corresponding to miRNA-221 cannot be detected. The merged image (yellow) confirms the generation of the green and red fluorescence by the tetrahedra. Figure 4, entry II, shows the bright-field microscopy image, the confocal fluorescence microscopy images at the three emission wavelengths, and the merge image of the HepG2 cells treated with the tetrahedra. Only the green fluorescence of FAM can be detected, and no Cy5 or ROX emission can be observed (merged green image). These results are consistent with the overexpression of miRNA-21 in the HepG2 cells. Figure 4, entry III, shows the bright-field microscopy image, the confocal fluorescence microscopy images at the three fluorophore imaging wavelengths, and the merge image of the MCF-10A cells treated with the tetrahedra. No fluorescence of the fluorophore probes associated with the tetrahedra is observed, consistent with the low expression of miRNA-21 or miRNA-155 in the MCF-10A cells (for large-area confocal fluorescence microscopy image of the different cells, see Figure S2, Supporting Information).
Figure 4.
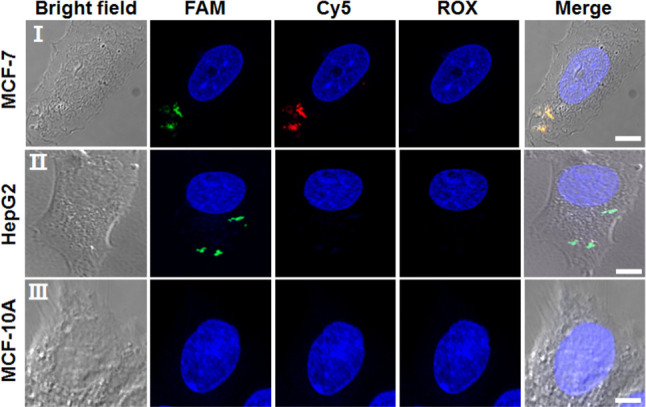
Bright-field microscopy images, confocal fluorescence microscopy images, and merge images corresponding to the analysis of (entry I) MCF-7 cells, (entry II) HepG2 cells, (entry III) MCF-10A cells treated with the FAM/Cy5/ROX-functionalized tetrahedra. The fluorescence of the three fluorescent probes is imaged through three channels: FAM emission, λex = 488 nm; Cy5 emission, λex = 640 nm; ROX emission, λex = 561 nm. Scale bar: 5 μm.
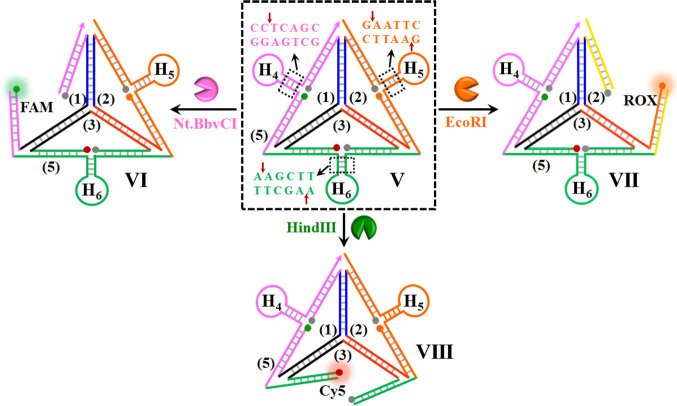
The tetrahedron sensing module appears to be a versatile sensing scaffold. By the appropriate engineering of the hairpin units associated with the module, other targets can be analyzed and detected in parallel. Scheme 2 presents the application of the tetrahedron scaffold to analyze enzymes that catalyze the cleavage of DNA. DNA cleavage by endonucleases is not only essential during cellular events and physiological processes, such as DNA replication or repair,59 but also extensively used as a powerful tool in PCR and molecular cloning.60 Thus, assaying of endonuclease activities is of significant importance in the fields of molecular biology and clinical diagnosis.61,62 As a proof of concept, three endonucleases, EcoRI, Nt.BbvCI, and HindIII, are analyzed in the present study. The sensing module includes, as before, the single strands 1, 2, and 3 functionalized with the different fluorophore/quencher pairs, and these are presented as three quasi-triangle structures exhibiting interhybridization domains. The three interhybridized quasi-triangles are rigidified by the peripheral strand 5 that includes, at the edges of the resulting tetrahedron, the hairpins H4, H5, and H6. The hairpins H4, H5, and H6 include duplex stem domains that are cleaved by the respective enzymes. In the rigidified tetrahedron structure, state V, all of the fluorophores exist in a quenched configuration. Subjecting the sensing module to Nt.BbvCI results in the cleavage of the stem region associated with hairpin H4. The cleavage of the hairpin associated with the respective edge results in a flexible edge, state VI, where the fluorophore FAM is spatially separated from the quencher unit. This leads to the switching-on fluorescence of FAM. Similarly, the treatment of the tetrahedron module with EcoRI leads to the cleavage of the stem domain of hairpin H5, and this transforms the H5-modified edge into a broken domain where the fluorophore ROX is spatially separated from the quencher, state VII. The flexibility of this edge is anticipated to switch on the luminescence of ROX. In addition, subjecting the sensing module to HindIII results in the base-specific nicking of one strand of the stem region of hairpin H6. The cleavage of the stem strand separates the hairpin and introduces functional flexibility into the H6 carrying edge. The separation of the rigidifying strand of this edge leads to a flexible structure of the domain, resulting in the spatial separation of the fluorophore Cy5 from the quencher unit, state VIII. As a result, the fluorescence of Cy5 is switched on.
Scheme 2. Schematic Multiplexed Analysis of Three Different Endonucleases Operating on a DNA Tetrahedra Module.
Figure 5A, panel I, shows the fluorescence spectra of the sensing module before interaction with Nt.BbvCI, curve (a), and after subjecting the sensing scaffold to Nt.BbvCI, 0.2 U μL–1, curve (b). Subjecting the sensing module to Nt.BbvCI leads to the triggered-on fluorescence of FAM, implying that hairpin H4 carrying edge was, indeed, cleaved, and the resulting flexibility and accompanied spatial separation of the fluorophore and quencher unit lead to the triggered-on fluorescence of FAM. Panel I, curves (c) and (d), show the effect of added HindIII or EcoRI catalyst, each at a concentration of 0.2 U μL–1, on the fluorescence of the fluorophore FAM. The fluorescence intensities of FAM are almost identical to the initial fluorescence intensity of FAM prior to the addition of Nt.BbvCI. Thus, although the addition of HindIII or EcoRI leads to structural transformation at other edges of the tetrahedron (vide infra), the fluorescence of FAM stays quenched in the presence of the other two biocatalysts. This indicates that the rigidified structure FAM/quencher is preserved, even though structural perturbations take place on remote edges associated with the tetrahedron structure. That is, the inner Y-shaped hybridization between the quasi-triangle subunits 1, 2, and 3 retains the basic skeleton, even though rigidified edges of the tetrahedron are broken. Figure 5A, panels II and III, show fluorescence changes of the sensing module upon its challenging with variable concentrations of Nt.BbvCI and the derived calibration curve, respectively. As the concentration of Nt.BbvCI increases, the fluorescence of FAM is intensified, consistent with the enhanced cleavage of the H4 carrying edge. Figure 5B, panel I, shows the fluorescence intensities of the fluorophore ROX, associated with the hairpin H5 carrying edge before treatment with EcoRI, curve (a), and after interaction with EcoRI, 0.16 U μL–1, curve (b). The addition of EcoRI results in the increased fluorescence of ROX, consistent with the EcoRI-stimulated cleavage of the hairpin H5 carrying edge and the accompanying spatial separation between the fluorophore ROX and quencher unit. Treatment of the sensing scaffold with Nt.BbvCI or HindIII does not affect the fluorescence of ROX, panel I, curves (c) and (d). Thus, although these biocatalysts induce structural transitions on the H4- and H6-hairpin-carrying edges, the quenched fluorescence of ROX is not affected. These results imply that the interhybridized Y-shaped structure of 1/2/3 stays intact, even though structural perturbations on the H4- or H6-haipin-carrying edges proceed. Figure 5B, panels II and III, show the fluorescence of ROX upon treatment of the sensing module with different concentrations of EcoRI and the derived calibration curve, respectively. The fluorescence intensities of ROX increase as the concentrations of EcoRI are elevated, consistent with the enhanced cleavage of the H5-carrying edge. The results demonstrate that the fluorescence changes of ROX are selective toward the sensing of EcoRI. Figure 5C, panel I, shows the fluorescence changes of the sensing module at fluorescence of Cy5 before its treatment with HindIII, curve (a), and after interaction of the sensing scaffold with the biocatalyst, 2.0 U μL–1, curve (b). The fluorescence of Cy5 is intensified, consistent with the cleavage of the tetrahedron edge carrying hairpin H6, state VIII. Subjecting the sensing module to the enzymes EcoRI or Nt.BbvCI does not affect the fluorescence of Cy5 curves (c) and (d). Thus, although the biocatalysts EcoRI and Nt.BbvCI are active in the cleavage of other domains, the fluorescence of Cy5 is not affected, implying that the fluorophore Cy5 is retained in its quenched configuration. Thus, beyond the selective sensing of HindIII, the results imply that the interhybridization of the Y-shaped domains of 1/2/3 retains their supramolecular structures. Figure 5C, panels II and III, show the fluorescence changes of the Cy5 reporter unit upon challenging the sensing module with different concentrations of HindIII and the resulting calibration curve, respectively. As the concentration of HindIII increases, the fluorescence of Cy5 is enhanced, consistent with higher degree of cleavage of the H6-carrying edge of the sensing module.
Figure 5.
Analysis of three endonucleases by the DNA tetrahedron module shown in Scheme 2. (A) Analysis of Nt.BbvCI. Panel I: fluorescence spectra of FAM (a) in the absence of added Nt.BbvCI, (b) upon the addition of Nt.BbvCI, 0.2 U μL–1, (c) upon addition of EcoRI, 0.2 U μL–1, (d) upon addition of HindIII, 0.2 U μL–1. Panel II: fluorescence changes of FAM upon the addition of variable concentrations of Nt.BbvCI. Panel III: derived calibration curve corresponding to the fluorescence changes of FAM at different concentrations of Nt.BbvCI. (B) Analysis of EcoRI. Panel I: fluorescence spectra of ROX (a) in the absence of added EcoRI, (b) upon the addition of EcoRI, 0.16 U μL–1, (c) upon addition of Nt.BbvCI, 0.16 U μL–1, (d) upon addition of HindIII, 0.16 U μL–1. Panel II: fluorescence changes of ROX upon the addition of variable concentrations of EcoRI. Panel III: derived calibration curve corresponding to the fluorescence changes of ROX at different concentrations of EcoRI. (C) Analysis of HindIII. Panel I: fluorescence spectra of Cy5 (a) in the absence of added HindIII, (b) upon the addition of HindIII, 2 U μL–1, (c) upon addition of Nt.BbvCI, 2 U μL–1, (d) upon addition of EcoRI, 2 U μL–1. Panel II: fluorescence changes of Cy5 upon the addition of variable concentrations of HindIII. Panel III: derived calibration curve corresponding to the fluorescence changes of Cy5 at different concentrations of HindIII.
The selective sensing of each of the enzyme-DNA-cleaving biocatalysts by the three-fluorophore-functionalized tetrahedron sensing module, Scheme 2, suggests that the multiplexed analysis of the enzymes should be feasible. Figure 6 shows the multiplexed analysis of the three enzymes by the tetrahedron module. Treatment of the sensing module with the two enzymes EcoRI and HindIII results in the fluorescence signals of the H5- and H6-hairpin-carrying edges corresponding to ROX and to Cy5. No fluorescence of FAM is detected, indicating that the H4 hairpin edge reporter is in the caged (quenched) state. Figure 6, panel II, depicts multiplexed analysis of EcoRI and Nt.BbvCI by the tetrahedra sensing platform. Treatment of the sensing module with the two enzymes yields the fluorescence of FAM and ROX, yet no fluorescence of Cy5 is observed, implying the H6-hairpin-caged fluorophore is retained upon sensing of the two enzymes. Also, Figure 6, panel III, shows the fluorescence bands of FAM and Cy5 upon sensing HindIII and Nt.BbvCI. No fluorescence of ROX is observed, indicating that the EcoRI-responsive H5-caged is retained under these conditions (fluorescence of ROX is quenched). Finally, panel IV depicts the fluorescence features of the tetrahedron sensing module in the presence of all three enzymes. All three fluorescence bands of FAM, ROX, and Cy5 are observed, demonstrating the successful multiplexed analysis of the three enzymes.
Figure 6.
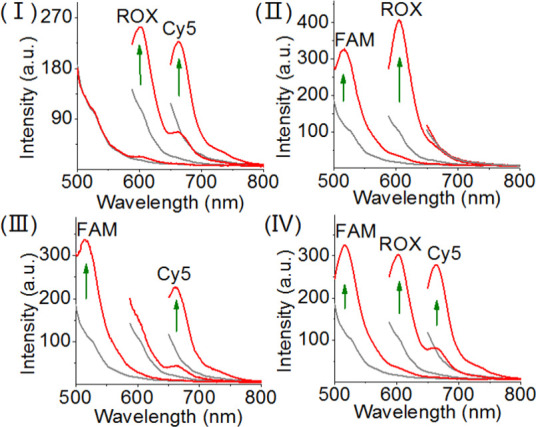
Multiplexed analysis of the three endonuclease, Nt.BbvCI, EcoRI, and HindIII by the DNA tetrahedron module shown in Scheme 2. Panel I: sensing of EcoRI and HindIII, switches on the fluorescence of ROX and Cy5. Panel II: sensing Nt.BbvCI and EcoRI by the sensing module, switches on the fluorescence of FAM and ROX. Panel III: sensing Nt.BbvCI and HindIII by the sensing module, switches on the fluorescence of FAM and Cy5. Panel IV: multiplexed sensing of all three endonucleases: Nt.BbvCI, EcoRI, and HindIII, switches on the fluorescence of FAM, ROX, and Cy5. In all experiments, the concentrations of Nt.BbvCI, EcoRI, and HindIII corresponded to 0.2, 0.2, and 2 U μL–1, respectively.
The generality of the tetrahedron module for the development of multiplexed sensing platforms was further demonstrated by the application of the tetrahedron scaffold for the multiplexed analysis of aptamer–ligands. Aptamers are sequence-specific nucleic acids generated by the SELEX (systematic evolution of ligands by exponential enrichment) process, which recognize low molecular weight substrates, proteins, and even cells.63,64 Different optical,65,66 electrochemical,67−69 and microgravimetric aptamer–ligand70 sensing platforms (aptasensors) were developed, and their bioanalytical and biomedical applications, particularly, multiplexed aptasensor configurations, were discussed.71,72Scheme 3 shows the development of tetrahedron sensing module for the parallel detection of an aptamer–ligand. The tetrahedron core consists of the triangle strands 1/2/3 and the long-strand 6 rigidifying the tetrahedron structure, state IX. The peripheral rigidifying strand 6 includes single-strand domains x, y, and z, and these are further hybridized with auxiliary strands x′, y′, and z′. This hybridization pattern yields a rigidified tetrahedron structure. Nonetheless, in contrast to the previous sensing modules that included fluorophore-quenched edges due to the proximity position of the fluorophore–quencher pairs, dictated by the peripheral rigidifying strand, in the present sensing module, the long and rigid duplex domains x/x′, y/y′, and z/z′ force the fluorophore–quencher pairs into spatially separated positions. Thus, the sensing module exhibits the selectively separated fluorescence bands of FAM, ROX, and Cy5, associated with the respective edges of the rigidified tetrahedron. The strands x′, y′, and z′ correspond to anti-ATP–aptamer, anti-VEGF–aptamer, and antithrombin aptamer, respectively. The parallel sensing principle of the different ligands is also detailed in the Scheme 3. In the presence of the ATP, the strand x′ is displaced in the form of the aptamer–ATP complex. The resulting single-strand x is pre-engineered to reconfigure after the displacement of x′ into the hairpin structure H7. The rigidification of the hairpin structure leads then to the forced proximity between the fluorophore and quencher on the H7-carrying edge, resulting in the fluorescence quenching of FAM, state X. Similarly, subjecting the sensing module to VEGF leads to the displacement of y′, through the formation of the aptamer–VEGF complex, state XI. This results in the reconfiguration of the single-strand domain into the hairpin H8. This leads to the close proximity between fluorophore (ROX) and the quencher, resulting in the quenching of the fluorophore. In addition, subjecting the sensing module to thrombin leads to the displacement of strand z′ through the formation of the aptamer–thrombin complex, state XII. The reconfiguration into the hairpin structure, H9, where the fluorophore (Cy5) and quencher are forced into close proximity that leads to effective quenching of the fluorophore, state XII. Note that the sensing of the respective ligands involves a negative fluorescence readout.
Scheme 3. Schematic Multiplexed Analysis of Three Aptamer–Ligands: Ligands = ATP, VEG,F and Thrombin by Tetrahedron Module (Note the Sensing Processes Are Accompanied by a Fluorescence Decrease, in Contrast to Previous Systems).
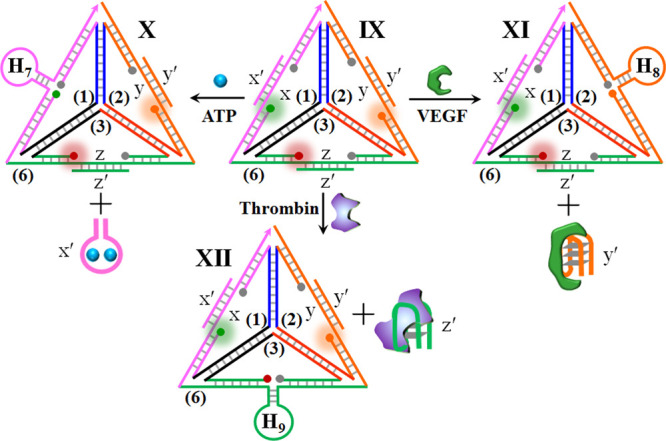
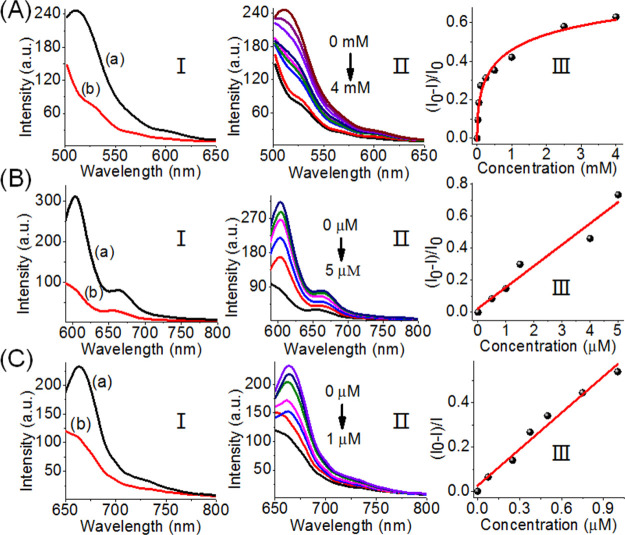
Figure 7 depicts the parallel analysis of the three ligands by the tetrahedron scaffold. Figure 7A, panel I, shows the fluorescence features of the sensing module at FAM fluorophore emission before addition of ATP, curve (a), and after the addition of ATP, 4 mM, curve (b). The fluorescence of FAM is quenched, consistent with the separation of the ATP–aptamer and the reconfiguration of the H7-carrying hairpin on the edge of the tetrahedron, state X. Figure 7A, panel II, shows the fluorescence intensities of FAM upon the addition of variable concentrations of ATP. As the concentrations of ATP increase, the quenching degree of the fluorescence of FAM is enhanced, consistent with the higher degree of the reconfigured H7-carrying edge, state X. It should be noted that the addition of either thrombin or VEGF to the tetrahedron sensing module does not have any effect on the fluorescence of FAM, even though their ligands have an effect on the reconfiguration of other domains of the tetrahedron module (see Figure S3). Figure 7B, panel I, shows the fluorescence of ROX associated with the sensing module before the addition of VEGF, curve (a), and after the addition of VEGF, 5 μM, curve (b). The fluorescence of ROX is quenched, consistent with the reconfiguration of the sensing module in state XI, where the dissociation of the anti-VEGF–aptamer resulted in the reconfiguration into the H8 tetrahedron carrying the H8-hairpin-modified edge. The addition of ATP or thrombin to the sensing module has no effect on the fluorescence of ROX, indicating that the fluorescence quenching is specific to the VEGF analyte (see Figure S3). Figure 7B, panel II, shows the gradual fluorescence changes of ROX upon increasing the concentrations of VEGF. As the concentrations of VEGF increase, the fluorescence quenching is enhanced. Figure 7B, panel III, reveals the derived calibration curve for the analysis of VEGF by the tetrahedron sensing module. Figure 7C, panel I, shows the fluorescence changes of Cy5 associated with the sensing module before treatment with thrombin, curve (a), and after treatment with thrombin, 1 μM, curve (b). The interaction of the sensing scaffold with thrombin leads to the quenching of Cy5, consistent with the displacement of the antithrombin aptamer from the module and the reconfiguration of the domain z into the hairpin H9, where the fluorophore Cy5 and the quencher units are forced into close proximity, state XII. The fluorescence of Cy5 is not affected by the addition of ATP or VEGF to the sensing scaffold, indicating that the observed changes in the fluorescence of Cy5 are specific to the formation of the thrombin–aptamer complex (see Figure S3). Figure 7C, panels II and III, show the fluorescence changes upon treatment of the sensing module with variable concentrations of thrombin and the derived calibration curve, respectively. The changes in the fluorescence intensities increase as the concentrations of thrombin are elevated.
Figure 7.
Analysis of three aptamer–ligands by the DNA tetrahedron module shown in Scheme 3. (A) Sensing of ATP by the DNA tetrahedron module. Panel I: fluorescence spectra of FAM before addition of ATP (a) and after the addition of ATP, 4 mM (b). Panel II: fluorescence changes of FAM upon interaction with different concentrations of ATP. Panel III: derived calibration curve corresponding to the fluorescence changes of FAM in the presence of variable concentrations of ATP. (B) Sensing of VEGF by the DNA tetrahedron module. Panel I: fluorescence spectra of ROX before addition of VEGF (a) and after the addition of VEGF, 5 μM (b). Panel II: fluorescence changes of ROX upon interaction with different concentrations of VEGF. Panel III: derived calibration curve corresponding to the fluorescence changes of ROX in the presence of variable concentrations of VEGF. (C) Sensing of thrombin by the DNA tetrahedron module. Panel I: fluorescence spectra of Cy5 before addition of thrombin (a) and after the addition of thrombin, 1 μM (b). Panel II: fluorescence changes of Cy5 upon interaction with different concentrations of thrombin. Panel III: derived calibration curve corresponding to the fluorescence changes of Cy5 in the presence of variable concentrations of thrombin.
The specific fluorescence changes transduced by the tetrahedron module in response to the respective ligands (thrombin, ATP, VEGF) were then applied to demonstrate the multiplexed analysis of the ligands (Figure 8). Treatment of the sensing scaffold with VEGF and thrombin results in the complete quenching of the fluorophore ROX and Cy5, whereas only a minute change in the fluorescence of FAM is observed (Figure 8, panel I). The results are consistent with the VEGF- and thrombin-driven displacement of the strand y′ and z′ in the form of the respective ligand–aptamer complexes and the reconfiguration of the hairpins H8 and H9 on the respective edges, leading to the effective quenching of ROX and Cy5. Under these conditions, the tetrahedron edge containing the x/x′ duplex is unaffected, resulting in the FAM fluorescence intensity characteristic to the parent tetrahedron. Similarly, subjecting the sensing module to the ligands ATP and thrombin leads to the quenching of the fluorescence of FAM and Cy5, whereas the fluorescence of ROX is unaffected. These results are consistent with the displacement of the strand x′ (by ATP) and of strand z′ (by thrombin) and the reconfiguration of the tetrahedron into the H7- and H9-hairpin-carrying edges. Also, Figure 8, panel III, shows the quenching fluorescence of FAM and ROX upon sensing ATP and VEGF. No fluorescence change of Cy5 is observed, indicating that the tetrahedron edge containing the z/z′ duplex is unaffected. Finally, the sensing module interacted with all ligand–analytes here (Figure 8, panel IV). All three fluorophore labels are quenched, demonstrating the multiplexed analysis of the three aptamer–ligand complexes.
Figure 8.
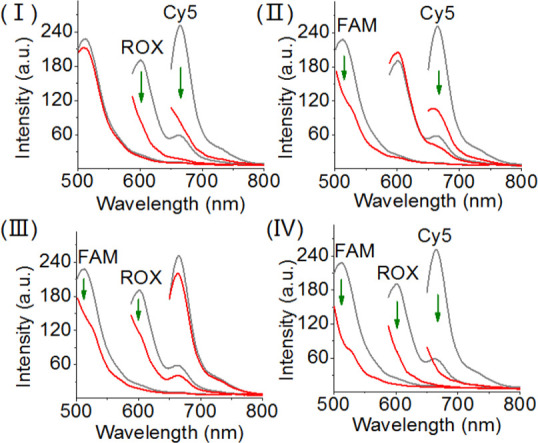
Multiplexed analysis of the three aptamer–ligands, ATP, VEGF, and thrombin by the DNA tetrahedron module shown in Scheme 3. Panel I: sensing of VEGF and thrombin, switches off the fluorescence of ROX and Cy5. Panel II: sensing ATP and thrombin by the sensing module, switches off the fluorescence of FAM and Cy5. Panel III: sensing ATP and VEGF by the sensing module, switches off the fluorescence of FAM and ROX. Panel IV: multiplexed sensing of all three aptamer–ligands: ATP, VEGF, and thrombin, switches off the fluorescence of FAM, ROX, and Cy5.
Conclusion
The study has introduced a versatile sensing module consisting of a DNA tetrahedron for the parallel and multiplexed analysis of different targets. In the present study, we engineered sensing modules for miRNAs, DNA-cleaving enzymes, and aptamer–ligands. These concepts can be, however, extended to other analytes, such as genes or metal ions. In the present study, we applied three fluorophores as the transducers of the parallel and multiplex analysis of the different analytes. By designing tetrahedra structures modified with other fluorophores and generating mixture of tetrahedra, the design of parallel and multiplexed sensing platforms of enhanced complexities may by envisaged. The facile permeation of the tetrahedra structure into cells was successfully applied to image MCF-7 and HepG2 cancer cells and their discrimination from the normal epithielial breast cell, MCF-10A, by a single sensing module.
Experimental Section
Preparation of the DNA Tetrahedron Module
For miRNA detection, four oligonucleotide strands including 1, 2, 3, and 4, 1 μM each was mixed in Tris-borate-EDTA (0.5× TBE) buffer containing 50 mM MgCl2, heated to 95 °C for 3 min, and then immediately cooled to 4 °C within 1 min. For enzyme detection, four oligonucleotide strands including 1, 2, 3, and 5, 1 μM each was mixed in 1× NEBuffer 2 (10 mM Tris, 20 mM MgCl2, 50 mM NaCl, 1 mM DTT and pH 7.9), heated to 95 °C for 3 min, and then immediately cooled to 4 °C within 1 min. For aptamer–ligand detection, seven oligonucleotide strands including 1 μM of 1, 2, 3, 6 and 1.5 μM of x′y′, and z′ were mixed in 1× NEBuffer 2 (10 mM Tris, 20 mM MgCl2, 50 mM NaCl, 1 mM DTT and pH 7.9), heated to 95 °C for 3 min, and then immediately cooled to 4 °C within 1 min.
Fluorescence Analysis
For detection of miRNAs, different concentrations of miRNAs were added to 100 μL of Tris-borate-EDTA (0.5× TBE) buffer solution (50 mM MgCl2) containing 100 nM DNA tetrahedron module, followed by incubation for 5 min at room temperature. For detection of enzymes, different concentrations of enzymes were added to 200 μL of 1× NEBuffer 2 (10 mM Tris, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT and pH 7.9), containing 90 nM DNA tetrahedron module, followed by incubation for 12 h at room temperature. For detection of aptamer–ligands, different concentrations of aptamer–ligands were added to 200 μL of 1× NEBuffer 2 (10 mM Tris, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT and pH 7.9), containing 90 nM DNA tetrahedron module, followed by incubation for 30 min at room temperature.
Cell Culture
Human breast cancer cells (MCF-7) were grown in 5% CO2 RPMI-1640 medium supplemented with 10% FCS, l-glutamine, and antibiotics (Biological Industries). Human liver cancer cell line (HepG2) were grown in 5% CO2 DMEM medium supplemented with 10% FCS, l-glutamine, and antibiotics (Biological Industries). Normal breast cells (MCF-10A) were maintained in complete growth medium consisting of 1:1 mixture of Dulbecco’s modified Eagle’s medium and Ham’s F12 medium supplemented with horse serum (5%), epidermal growth factor (20 ng/mL), cholera toxin (CT, 0.1 μg/mg), insulin (10 μg/mL), hydrocortisone (500 ng/mL), and penicillin/streptomycin (1 unit/mL). Cells were plated 1 day prior to the experiment on a μ-slide 4-well glass bottom dish (ibidi) for confocal microscopy.
Confocal Microscopy Measurements
For cell imaging experiments, cells were planted on a μ-slide 4-well glass bottom dish (ibidi). Cells were incubated with the DNA tetrahedron nanostructures after being washed with phosphate-buffered saline. The three samples of DNA tetrahedra (300 nM) were incubated with cells for 5 h and then washed with DMEM/Hepes twice and replenished with the fresh medium for the measurement. The fluorescence of tetrahedra in cells was monitored with an Olympus FV3000 confocal laser scanning microscope, and all images were analyzed with ImageJ.
Acknowledgments
This study is supported by the Israel Science Foundation and the Minerva Center for Biohybrid Complex Systems.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.0c04031.
Nucleic acid sequences, materials, measurement methods, agarose gel for multiplex detection of miRNAs, large-area confocal fluorescence images, and additional fluorescence spectra of aptamer–ligand detection (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Han M.; Gao X.; Su J. Z.; Nie S. Quantum-Dot-Tagged Microbeads for Multiplexed Optical Coding of Biomolecules. Nat. Biotechnol. 2001, 19, 631–635. 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- Lockhart D. J.; Winzeler E. A. Genomics, Gene Expression and DNA Arrays. Nature 2000, 405, 827–836. 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- Xiao M.; Lai W.; Man T.; Chang B.; Li L.; Chandrasekaran A. R.; Pei H. Rationally Engineered Nucleic Acid Architectures for Biosensing Applications. Chem. Rev. 2019, 119, 11631–11717. 10.1021/acs.chemrev.9b00121. [DOI] [PubMed] [Google Scholar]
- Stoeva S. I.; Lee J.-S.; Thaxton C. S.; Mirkin C. A. Multiplexed DNA Detection with Biobarcoded Nanoparticle Probes. Angew. Chem., Int. Ed. 2006, 45, 3303–3306. 10.1002/anie.200600124. [DOI] [PubMed] [Google Scholar]
- Heller M. J. DNA Microarray Technology: Devices, Systems, and Applications. Annu. Rev. Biomed. Eng. 2002, 4, 129–153. 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- Liu J.; Cao Z.; Lu Y. Functional Nucleic Acid Sensors. Chem. Rev. 2009, 109, 1948–1998. 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri S.; Sykes E. A.; Jennings T. L.; Chan W. C. W. Rapid Screening of Genetic Biomarkers of Infectious Agents Using Quantum Dot Barcodes. ACS Nano 2011, 5, 1580–1587. 10.1021/nn102873w. [DOI] [PubMed] [Google Scholar]
- Song L.; Ahn S.; Walt D. R. Fiber-Optic Microsphere-Based Arrays for Multiplexed Biological Warfare Agent Detection. Anal. Chem. 2006, 78, 1023–1033. 10.1021/ac051417w. [DOI] [PubMed] [Google Scholar]
- Nicewarner-Pena S. R.; Freeman R. G.; Reiss B. D.; He L.; Peña D. J.; Walton I. D.; Cromer R.; Keating C. D.; Natan M. J. Submicrometer Metallic Barcodes. Science 2001, 294, 137–141. 10.1126/science.294.5540.137. [DOI] [PubMed] [Google Scholar]
- Liu X.; Aizen R.; Freeman R.; Yehezkeli O.; Willner I. Multiplexed Aptasensors and Amplified DNA Sensors Using Functionalized Graphene Oxide: Application for Logic Gate Operations. ACS Nano 2012, 6, 3553–3563. 10.1021/nn300598q. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Zheng B.; Zhu C.; Zhang X.; Tan C.; Li H.; Chen B.; Yang J.; Chen J.; Huang Y.; Wang L.; Zhang H. Single-Layer Transition Metal Dichalcogenide Nanosheet-Based Nanosensors for Rapid, Sensitive, and Multiplexed Detection of DNA. Adv. Mater. 2015, 27, 935–939. 10.1002/adma.201404568. [DOI] [PubMed] [Google Scholar]
- Gill R.; Zayats M.; Willner I. Semiconductor Quantum Dots for Bioanalysis. Angew. Chem., Int. Ed. 2008, 47, 7602–7625. 10.1002/anie.200800169. [DOI] [PubMed] [Google Scholar]
- Freeman R.; Liu X.; Willner I. Amplified Multiplexed Analysis of DNA by the Exonuclease III-Catalyzed Regeneration of the Target DNA in the Presence of Functionalized Semiconductor Quantum Dots. Nano Lett. 2011, 11, 4456–4461. 10.1021/nl202761g. [DOI] [PubMed] [Google Scholar]
- Enkin N.; Wang F.; Sharon E.; Albada H. B.; Willner I. Multiplexed Analysis of Genes Using Nucleic Acid-Stabilized Silver-Nanocluster Quantum Dots. ACS Nano 2014, 8, 11666–11673. 10.1021/nn504983j. [DOI] [PubMed] [Google Scholar]
- Enkin N.; Sharon E.; Golub E.; Willner I. Ag Nanocluster/DNA Hybrids: Functional Modules for the Detection of Nitroaromatic and RDX Explosives. Nano Lett. 2014, 14, 4918–4922. 10.1021/nl502720s. [DOI] [PubMed] [Google Scholar]
- Freeman R.; Liu X.; Willner I. Chemiluminescent and Chemiluminescence Resonance Energy Transfer (CRET) Detection of DNA, Metal Ions, and Aptamer–Substrate Complexes Using Hemin/G-Quadruplexes and CdSe/ZnS Quantum Dots. J. Am. Chem. Soc. 2011, 133, 11597–11604. 10.1021/ja202639m. [DOI] [PubMed] [Google Scholar]
- Liu X.; Freeman R.; Golub E.; Willner I. Chemiluminescence and Chemiluminescence Resonance Energy Transfer (CRET) Aptamer Sensors Using Catalytic Hemin/G-Quadruplexes. ACS Nano 2011, 5, 7648–7655. 10.1021/nn202799d. [DOI] [PubMed] [Google Scholar]
- Hansen J. A.; Wang J.; Kawde A.-N.; Xiang Y.; Gothelf K. V.; Collins G. Quantum-Dot/Aptamer-Based Ultrasensitive Multi-Analyte Electrochemical Biosensor. J. Am. Chem. Soc. 2006, 128, 2228–2229. 10.1021/ja060005h. [DOI] [PubMed] [Google Scholar]
- Hildebrandt N. Biofunctional Quantum Dots: Controlled Conjugation for Multiplexed Biosensors. ACS Nano 2011, 5, 5286–5290. 10.1021/nn2023123. [DOI] [PubMed] [Google Scholar]
- Wang J.; Liu G.; Merkoçi A. Electrochemical Coding Technology for Simultaneous Detection of Multiple DNA Targets. J. Am. Chem. Soc. 2003, 125, 3214–3215. 10.1021/ja029668z. [DOI] [PubMed] [Google Scholar]
- Du Y.; Chen C.; Zhou M.; Dong S.; Wang E. Microfluidic Electrochemical Aptameric Assay Integrated On-Chip: A Potentially Convenient Sensing Platform for the Amplified and Multiplex Analysis of Small Molecules. Anal. Chem. 2011, 83, 1523–1529. 10.1021/ac101988n. [DOI] [PubMed] [Google Scholar]
- Wang F.; Elbaz J.; Orbach R.; Magen N.; Willner I. Amplified Analysis of DNA by the Autonomous Assembly of Polymers Consisting of DNAzyme Wires. J. Am. Chem. Soc. 2011, 133, 17149–17151. 10.1021/ja2076789. [DOI] [PubMed] [Google Scholar]
- Tian Y.; He Y.; Mao C. Cascade Signal Amplification for DNA Detection. ChemBioChem 2006, 7, 1862–1864. 10.1002/cbic.200600336. [DOI] [PubMed] [Google Scholar]
- Wang F.; Lu C.-H.; Liu X.; Freage L.; Willner I. Amplified and Multiplexed Detection of DNA Using the Dendritic Rolling Circle Amplified Synthesis of DNAzyme Reporter Units. Anal. Chem. 2014, 86, 1614–1621. 10.1021/ac4033033. [DOI] [PubMed] [Google Scholar]
- Wang F.; Freage L.; Orbach R.; Willner I. Autonomous Replication of Nucleic Acids by Polymerization/Nicking Enzyme/DNAzyme Cascades for the Amplified Detection of DNA and the Aptamer–Cocaine Complex. Anal. Chem. 2013, 85, 8196–8203. 10.1021/ac4013094. [DOI] [PubMed] [Google Scholar]
- Seeman N. C.; Sleiman H. F. DNA Nanotechnology. Nat. Rev. Mater. 2018, 3, 17068. 10.1038/natrevmats.2017.68. [DOI] [Google Scholar]
- Lu C.-H.; Willner B.; Willner I. DNA Nanotechnology: From Sensing and DNA Machines to Drug-Delivery Systems. ACS Nano 2013, 7, 8320–8332. 10.1021/nn404613v. [DOI] [PubMed] [Google Scholar]
- Chen C.; Geng J.; Pu F.; Yang X.; Ren J.; Qu X. Polyvalent Nucleic Acid/Mesoporous Silica Nanoparticle Conjugates: Dual Stimuli-Responsive Vehicles for Intracellular Drug Delivery. Angew. Chem., Int. Ed. 2011, 50, 882–886. 10.1002/anie.201005471. [DOI] [PubMed] [Google Scholar]
- Zhang P.; Cheng F.; Zhou R.; Cao J.; Li J.; Burda C.; Min Q.; Zhu J.-J. DNA-Hybrid-Gated Multifunctional Mesoporous Silica Nanocarriers for Dual-Targeted and MicroRNA-Responsive Controlled Drug Delivery. Angew. Chem., Int. Ed. 2014, 53, 2371–2375. 10.1002/anie.201308920. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Balogh D.; Wang F.; Willner I. Smart Mesoporous SiO2 Nanoparticles for the DNAzyme-Induced Multiplexed Release of Substrates. J. Am. Chem. Soc. 2013, 135, 1934–1940. 10.1021/ja311385y. [DOI] [PubMed] [Google Scholar]
- Lu C.-H.; Willner I. Stimuli-Responsive DNA-Functionalized Nano-/Microcontainers for Switchable and Controlled Release. Angew. Chem., Int. Ed. 2015, 54, 12212–12235. 10.1002/anie.201503054. [DOI] [PubMed] [Google Scholar]
- Chen W.-H.; Liao W.-C.; Sohn Y. S.; Fadeev M.; Cecconello A.; Nechushtai R.; Willner I. Stimuli-Responsive Nucleic Acid-Based Polyacrylamide Hydrogel-Coated Metal-Organic Framework Nanoparticles for Controlled Drug Release. Adv. Funct. Mater. 2018, 28, 1705137. 10.1002/adfm.201705137. [DOI] [Google Scholar]
- Chen W.-H.; Luo G.-F.; Vázquez-González M.; Cazelles R.; Sohn Y. S.; Nechushtai R.; Mandel Y.; Willner I. Glucose-Responsive Metal–Organic-Framework Nanoparticles Act as “Smart” Sense-and-Treat Carriers. ACS Nano 2018, 12, 7538–7545. 10.1021/acsnano.8b03417. [DOI] [PubMed] [Google Scholar]
- Chen W.-H.; Yu X.; Cecconello A.; Sohn Y. S.; Nechushtai R.; Willner I. Stimuli-Responsive Nucleic Acid-Functionalized Metal–Organic Framework Nanoparticles Using pH- and Metal-Ion-Dependent DNAzymes as Locks. Chem. Sci. 2017, 8, 5769–5780. 10.1039/C7SC01765K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-H.; Yu X.; Liao W.-C.; Sohn Y. S.; Cecconello A.; Kozell A.; Nechushtai R.; Willner I. ATP-Responsive Aptamer-Based Metal-Organic Framework Nanoparticles (NMOFs) for the Controlled Release of Loads and Drugs. Adv. Funct. Mater. 2017, 27, 1702102. 10.1002/adfm.201702102. [DOI] [Google Scholar]
- Liao W.-C.; Riutin M.; Parak W. J.; Willner I. Programmed pH-Responsive Microcapsules for the Controlled Release of CdSe/ZnS Quantum Dots. ACS Nano 2016, 10, 8683–8689. 10.1021/acsnano.6b04056. [DOI] [PubMed] [Google Scholar]
- Liao W.-C.; Sohn Y. S.; Riutin M.; Cecconello A.; Parak W. J.; Nechushtai R.; Willner I. The Application of Stimuli-Responsive VEGF- and ATP-Aptamer-Based Microcapsules for the Controlled Release of an Anticancer Drug, and the Selective Targeted Cytotoxicity toward Cancer Cells. Adv. Funct. Mater. 2016, 26, 4262–4273. 10.1002/adfm.201600069. [DOI] [Google Scholar]
- Pinheiro A. V.; Han D.; Shih W. M.; Yan H. Challenges and Opportunities for Structural DNA Nanotechnology. Nat. Nanotechnol. 2011, 6, 763–772. 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E. S.; Dong M.; Nielsen M. M.; Jahn K.; Subramani R.; Mamdouh W.; Golas M. M.; Sander B.; Stark H.; Oliveira C. L. P.; Pedersen J. S.; Birkedal V.; Besenbacher F.; Gothelf K. V.; Kjems J. Self-Assembly of a Nanoscale DNA Box with a Controllable Lid. Nature 2009, 459, 73–76. 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- Wang X.; Chandrasekaran A. R.; Shen Z.; Ohayon Y. P.; Wang T.; Kizer M. E.; Sha R.; Mao C.; Yan H.; Zhang X.; Liao S.; Ding B.; Chakraborty B.; Jonoska N.; Niu D.; Gu H.; Chao J.; Gao X.; Li Y.; Ciengshin T.; Seeman N. C. Paranemic Crossover DNA: There and Back Again. Chem. Rev. 2019, 119, 6273–6289. 10.1021/acs.chemrev.8b00207. [DOI] [PubMed] [Google Scholar]
- Mao C.; Sun W.; Seeman N. C. Designed Two-Dimensional DNA Holliday Junction Arrays Visualized by Atomic Force Microscopy. J. Am. Chem. Soc. 1999, 121, 5437–5443. 10.1021/ja9900398. [DOI] [Google Scholar]
- Goodman R. P.; Schaap I. A. T.; Tardin C. F.; Erben C. M.; Berry R. M.; Schmidt C. F.; Turberfield A. J. Rapid Chiral Assembly of Rigid DNA Building Blocks for Molecular Nanofabrication. Science 2005, 310, 1661–1665. 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- Goodman R. P.; Berry R. M.; Turberfield A. J. The Single-Step Synthesis of a DNA Tetrahedron. Chem. Commun. 2004, 1372. 10.1039/b402293a. [DOI] [PubMed] [Google Scholar]
- Goodman R. P.; Heilemann M.; Doose S.; Erben C. M.; Kapanidis A. N.; Turberfield A. J. Reconfigurable, Braced, Three-Dimensional DNA Nanostructures. Nat. Nanotechnol. 2008, 3, 93–96. 10.1038/nnano.2008.3. [DOI] [PubMed] [Google Scholar]
- Zhou Z.; Zhang P.; Yue L.; Willner I. Triggered Interconversion of Dynamic Networks Composed of DNA-Tetrahedra Nanostructures. Nano Lett. 2019, 19, 7540–7547. 10.1021/acs.nanolett.9b03606. [DOI] [PubMed] [Google Scholar]
- Pei H.; Lu N.; Wen Y.; Song S.; Liu Y.; Yan H.; Fan C. A DNA Nanostructure-Based Biomolecular Probe Carrier Platform for Electrochemical Biosensing. Adv. Mater. 2010, 22, 4754–4758. 10.1002/adma.201002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh A. S.; Yin H.; Erben C. M.; Wood M. J. A.; Turberfield A. J. DNA Cage Delivery to Mammalian Cells. ACS Nano 2011, 5, 5427–5432. 10.1021/nn2005574. [DOI] [PubMed] [Google Scholar]
- Li J.; Pei H.; Zhu B.; Liang L.; Wei M.; He Y.; Chen N.; Li D.; Huang Q.; Fan C. Self-Assembled Multivalent DNA Nanostructures for Noninvasive Intracellular Delivery of Immunostimulatory CpG Oligonucleotides. ACS Nano 2011, 5, 8783–8789. 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- Peng P.; Du Y.; Zheng J.; Wang H.; Li T. Reconfigurable Bioinspired Framework Nucleic Acid Nanoplatform Dynamically Manipulated in Living Cells for Subcellular Imaging. Angew. Chem., Int. Ed. 2019, 58, 1648–1653. 10.1002/anie.201811117. [DOI] [PubMed] [Google Scholar]
- Pei H.; Liang L.; Yao G.; Li J.; Huang Q.; Fan C. Reconfigurable Three-Dimensional DNA Nanostructures for the Construction of Intracellular Logic Sensors. Angew. Chem., Int. Ed. 2012, 51, 9020–9024. 10.1002/anie.201202356. [DOI] [PubMed] [Google Scholar]
- He L.; Lu D.; Liang H.; Xie S.; Zhang X.; Liu Q.; Yuan Q.; Tan W. mRNA-Initiated, Three-Dimensional DNA Amplifier Able to Function inside Living Cells. J. Am. Chem. Soc. 2018, 140, 258–263. 10.1021/jacs.7b09789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.; MicroRNAs P. Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres S. L.; Horwich M. D.; Hung J. H.; Xu J.; Ghildiyal M.; Weng Z.; Zamore P. D. Target RNA-Directed Trimming and Tailing of Small Silencing RNAs. Science 2010, 328, 1534–1539. 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton A. E.; Gall T. M. H.; Castellano L.; Stebbing J.; Jiao L. R.; Krell J. Towards a Clinical Use of miRNAs in Pancreatic Cancer Biopsies. Expert Rev. Mol. Diagn. 2013, 13, 31–34. 10.1586/erm.12.136. [DOI] [PubMed] [Google Scholar]
- Ma R.; Jiang T.; Kang X. Circulating MicroRNAs in Cancer: Origin, Function and Application. J. Exp. Clin. Cancer Res. 2012, 31, 38. 10.1186/1756-9966-31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H.; Zhang J.; Ju H.; Lu H.; Wang S.; Jin S.; Hao K.; Du H.; Zhang X. Highly Sensitive Multiple MicroRNA Detection Based on Fluorescence Quenching of Graphene Oxide and Isothermal Strand-Displacement Polymerase Reaction. Anal. Chem. 2012, 84, 4587–4593. 10.1021/ac300721u. [DOI] [PubMed] [Google Scholar]
- Gong X.; Wei J.; Liu J.; Li R.; Liu X.; Wang F. Programmable Intracellular DNA Biocomputing Circuits for Reliable Cell Recognitions. Chem. Sci. 2019, 10, 2989–2997. 10.1039/C8SC05217D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J.; Wu F.; Xia H.; Wu Y.; Liu S. Accurate Cancer Cell Identification and MicroRNA Silencing Induced Therapy Using Tailored DNA Tetrahedron Nanostructures. Chem. Sci. 2020, 11, 80–86. 10.1039/C9SC04823E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction Enzymes and Their Isoschizomers. Nucleic Acids Res. 1990, 18, 2331–2365. 10.1093/nar/18.suppl.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M.; Benimetskaya L.; Lebedeva I.; Dignam J.; Takle G.; Stein C. A. Intracellular mRNA Cleavage Induced through Activation of RNase P by Nuclease-Resistant External Guide Sequences. Nat. Biotechnol. 2000, 18, 58–61. 10.1038/71924. [DOI] [PubMed] [Google Scholar]
- Feng X.; Duan X.; Liu L.; Feng F.; Wang S.; Li Y.; Zhu D. Fluorescence Logic-Signal-Based Multiplex Detection of Nucleases with the Assembly of a Cationic Conjugated Polymer and Branched DNA. Angew. Chem., Int. Ed. 2009, 48, 5316–5321. 10.1002/anie.200901555. [DOI] [PubMed] [Google Scholar]
- Hu J.; Liu M.-H.; Zhang C.-Y. Construction of Tetrahedral DNA-Quantum Dot Nanostructure with the Integration of Multistep Förster Resonance Energy Transfer for Multiplex Enzymes Assay. ACS Nano 2019, 13, 7191–7201. 10.1021/acsnano.9b02679. [DOI] [PubMed] [Google Scholar]
- Jayasena S.; Aptamers D. An Emerging Class of Molecules that Rival Antibodies in Diagnostics. Clin. Chem. 1999, 45, 1628–1650. 10.1093/clinchem/45.9.1628. [DOI] [PubMed] [Google Scholar]
- Osborne S. E.; Matsumura I.; Ellington A. D. Aptamers as Therapeutic and Diagnostic Reagents: Problems and Prospects. Curr. Opin. Chem. Biol. 1997, 1, 5–9. 10.1016/S1367-5931(97)80102-0. [DOI] [PubMed] [Google Scholar]
- Ho H.-A.; Leclerc M. Optical Sensors Based on Hybrid Aptamer/Conjugated Polymer Complexes. J. Am. Chem. Soc. 2004, 126, 1384–1387. 10.1021/ja037289f. [DOI] [PubMed] [Google Scholar]
- Canoura J.; Wang Z.; Yu H.; Alkhamis O.; Fu F.; Xiao Y. No Structure-Switching Required: A Generalizable Exonuclease-Mediated Aptamer-Based Assay for Small-Molecule Detection. J. Am. Chem. Soc. 2018, 140, 9961–9971. 10.1021/jacs.8b04975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y.; Pei H.; Wan Y.; Su Y.; Huang Q.; Song S.; Fan C. DNA Nanostructure-Decorated Surfaces for Enhanced Aptamer-Target Binding and Electrochemical Cocaine Sensors. Anal. Chem. 2011, 83, 7418–7423. 10.1021/ac201491p. [DOI] [PubMed] [Google Scholar]
- Xiao Y.; Lubin A. A.; Heeger A. J.; Plaxco K. W. Label-Free Electronic Detection of Thrombin in Blood Serum by Using an Aptamer-Based Sensor. Angew. Chem., Int. Ed. 2005, 44, 5456–5459. 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- Willner I.; Zayats M. Electronic Aptamer-Based Sensors. Angew. Chem., Int. Ed. 2007, 46, 6408–6418. 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- Pavlov V.; Xiao Y.; Shlyahovsky B.; Willner I. Aptamer-Functionalized Au Nanoparticles for the Amplified Optical Detection of Thrombin. J. Am. Chem. Soc. 2004, 126, 11768–11769. 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- Famulok M.; Mayer G. Aptamer Modules as Sensors and Detectors. Acc. Chem. Res. 2011, 44, 1349–1358. 10.1021/ar2000293. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Wang L.; Zhang H.; Boey F.; Song S.; Fan C. Aptamer-Based Multicolor Fluorescent Gold Nanoprobes for Multiplex Detection in Homogeneous Solution. Small 2010, 6, 201–204. 10.1002/smll.200901012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.