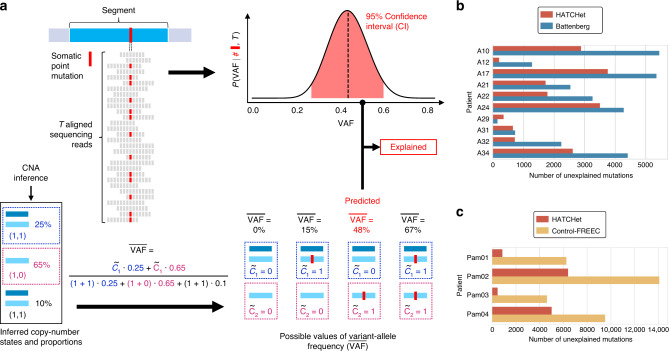
Fig. 6. HATCHet infers copy-number states and proportions that better explain VAFs of somatic SNVs and small indels.
a A genomic segment (cyan rectangle) harbors a somatic mutation, which corresponds to either a somatic SNV or small indel. Reads with variant allele (red squares) and reference allele (gray squares) are used to estimate the VAF. (Top right) From T sequencing reads (gray rectangles) covering the mutation, a 95% confidence interval (CI, i.e., red area of posterior probability) on the VAF is obtained from a binomial model. (Bottom) Separately, copy-number states and proportions are inferred for this genomic segment. Given the numbers of mutated copies in each of the two copy-number states, the of the mutation is computed as the fraction of the mutated copies weighted by the proportions of the corresponding copy-number states. Assuming that an allele-specific position is mutated at most once during tumor progression (i.e., no homoplasy), all possible values of are computed according to the possible values of and . A mutation is explained if at least one value of is within CI. b Over 10,600 mutations identified per prostate cancer patient on average, HATCHet copy numbers (red) yield fewer unexplained mutations than Battenberg (blue) in all patients but A29, where the difference is small. c Over 9,000 mutations identified per pancreas cancer patient on average, HATCHet copy numbers yield fewer unexplained mutations in all patients than Control-FREEC.