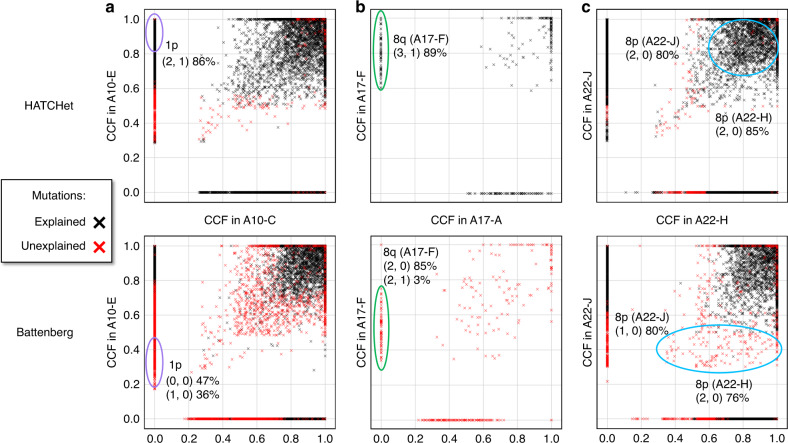
Fig. 7. HATCHet copy numbers improve estimates of CCFs of somatic mutations in prostate cancer patients.
a CCFs of somatic SNVs and small indels in samples A10-C and A10-E of patient A10 computed from allele-specific copy numbers and proportions inferred by HATCHet (top) and Battenberg (bottom). HATCHet explains a substantial number of mutations that are unexplained by Battenberg; for example, HATCHet infers a clonal CNA on chromosome 1p in A10-E and determines that the mutations at this locus (purple circle) are clonal (i.e., CCF ≈ 1). In contrast, Battenberg infers subclonal CNAs at the same locus, and determines that the same mutations are subclonal (CCF ≈ 0.3). b CCFs of somatic SNVs and small indels in samples A17-A and A17-F of patient A17 show groups of mutations that are explained by HATCHet and unexplained by Battenberg (only this subset of mutations is shown here for simplicity). For example, HATCHet infers a clonal CNA on chromosome 8q in A17-F and suggests that mutations in that region (green circle) are clonal (CCF ≈ 1), while Battenberg infers subclonal CNAs and suggests that the same mutations are subclonal (CCF ≈ 0.5). c CCFs of somatic SNVs and small indels in samples A22-J and A22-H of patient A22 show a large group of shared mutations on chromosome 8p (cyan circle with CCF > 0 in both samples). HATCHet infers the same copy-number state (2, 0) in both samples, explains these mutations, and suggests that they are clonal. Battenberg infers distinct copy-number states (1, 0) and (2, 0) in the two samples, leaves these mutations unexplained, and suggests that the mutations are subclonal in both samples.