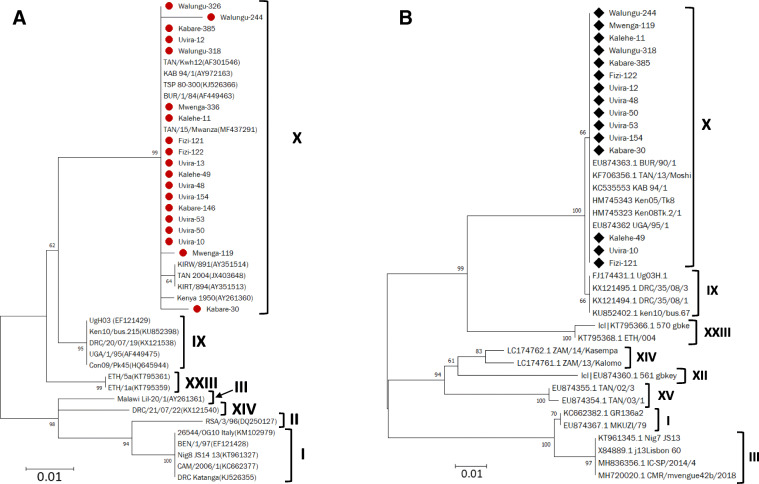
Fig. 2.
Phylogenetic relationships of p72 and p54 genotypes. The evolutionary history was inferred by the maximum likelihood method based on the Kimura 2-parameter model [33]. Phylogeny was inferred following 1000 bootstrap replications, and the node values show percentage bootstrap support. Scale bar indicates nucleotide substitutions per site. Scale bar indicates nucleotide substitutions per site. a p72 genotypes. The analysis included 19 B646L (p72) sequences from this study (plain circle  ) and sequences from the GenBank database. The GenBank accession numbers for the different B646L (p72) genes are indicated in parenthesis. b p54 genotypes. The analysis involved 15 E183L (p54) gene sequences of African swine fever viruses from this study (black diamond ◆) and sequences from the GenBank database. The sequences for the different B646L (p72) and E183L (p54) genes are starting by GenBank accession numbers
) and sequences from the GenBank database. The GenBank accession numbers for the different B646L (p72) genes are indicated in parenthesis. b p54 genotypes. The analysis involved 15 E183L (p54) gene sequences of African swine fever viruses from this study (black diamond ◆) and sequences from the GenBank database. The sequences for the different B646L (p72) and E183L (p54) genes are starting by GenBank accession numbers