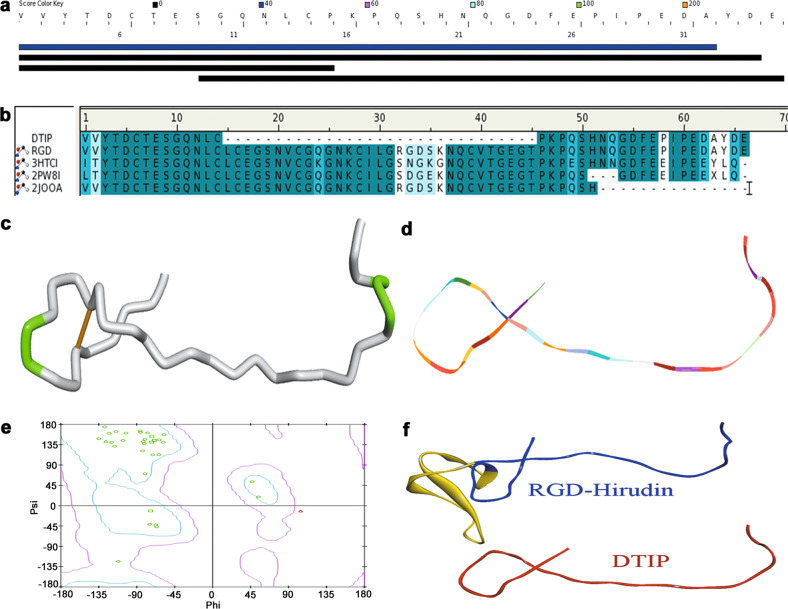
Fig. 3. Molecule simulation of DTIP.
a Similar sequences with DTIP in DS server database. b Amino acid sequence alignment of DTIP with RGD-hirudin, recombinant hirudin, sulfo-hirudin, and recombinant RGD-hirudin. Same amino acid residues are shown in dark blue, while similar amino acid residues are shown in light blue. c Homology modeling of DTIP in a tube format. d Homology modeling of DTIP in a line ribbon format. e Ramachandran Plot of the DTIP model. The distribution of the DTIP residues (green ring) are shown in colors: allowed region (within light blue), marginal region (beyond light blue and within pink), and disallowed region (beyond pink). The percent of residues in the allowed region was 96.3%, in the marginal region 3.7%, and in the disallowed region 0.0%. f Homology modeling of RGD-hirudin and DTIP in a flat ribbon format (blue and red, respectively) and the fragment from Leu15 to Thr45 (yellow).