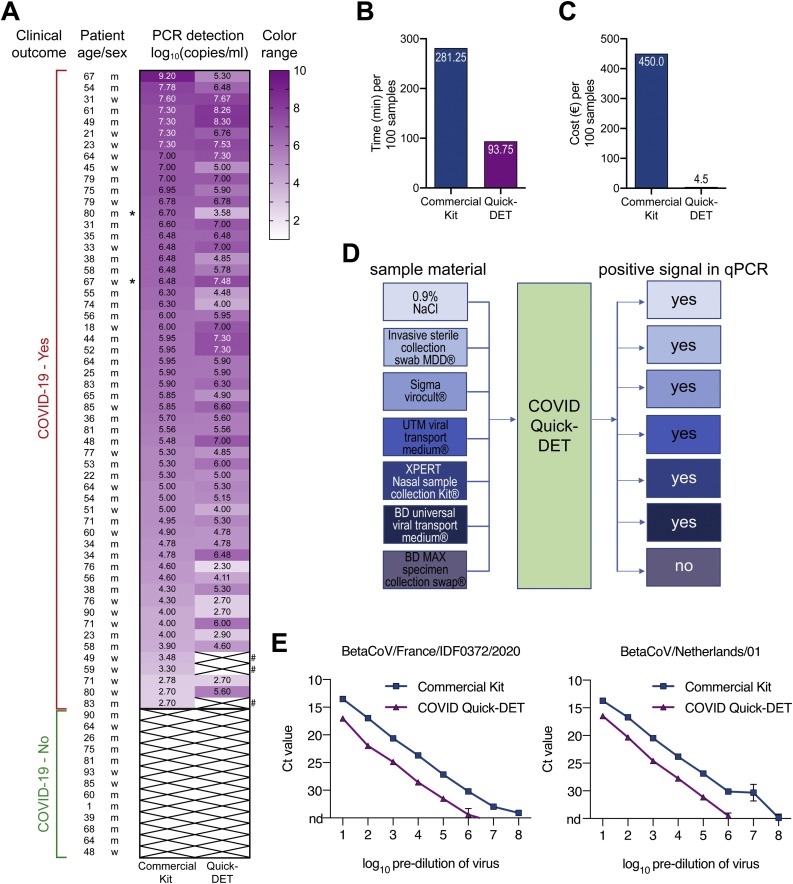
Fig. 1.
(A) Heat map of SARS-CoV-2 gene equivalents (log10) for 56 COVID-19 patients using a commercial RNA isolation kit or COVID-quick-DET. The colors in the heat map represent gene expression levels (log10). White crossed boxes marked with “#” represent the samples giving discrepant results between COVID-quick-DET and the reference method. Samples taken from bronchoalveolar lavages are indicated by asterisk. (B) Time for RNA isolation per 100 samples in minutes. (C) Costs for RNA isolation per 100 samples in Euro. (D) Schematic workflow of tested material compatible with COVID-Quick-DET method and successful qPCR. (E) Detection of SARS-CoV-2 ORF1b-nsp14 in samples prepared by serial dilution of virus stocks of two strains in medium (French-left panel; Dutch-right panel). Viral RNA was either extracted by Qiagen Viral RNA Mini Kit (Qiagen #52906) or prepared by the COVID-quick-DET protocol before being subjected to RT-qPCR as described (Chu et al., 2020; https://www.who.int/). For direct comparison to COVID-quick-DET, RNA was eluted in the original sample volume without concentration using Qiagen Viral RNA Mini Kit. Ct 35 was set as cut-off. Reactions have been run in duplicates, values are mean ± SD. N.d. = not detected (Ct => 35).