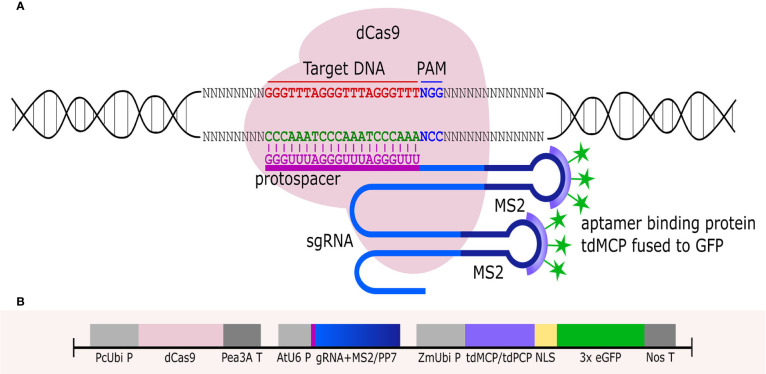
Figure 1.
RNA aptamer-based CRISPR/dCas9 imaging of telomere repeats. (A) Schemata depicting the components of the aptamer-based CRISPR labeling method: (1) dCas9 from S. pyogenes, (2) MS2 or PP7 aptamers (here only MS2 is shown) which are integrated into the sgRNA scaffold. (3) RNA binding protein (tdMCP or tdPCP) fused to fluorescent protein (3x eGFP) which recognizes aptamers. Protospacer designed to target Arabidopsis-type telomere DNA sequence. (B) Structure of the aptamer-based CRISPR imaging construct. dCas9 is driven by a ubiquitin promoter from parsley (PcUbi P), chimeric gRNA including aptamers (MS2/PP7) are driven by the AtU6 promoter (AtU6 P), aptamer binding proteins fused to a fluorescent protein (tdMCP/tdPCP) with the help of nuclear localization signal (NLS) are driven by a ubiquitin promoter from maize (ZmUbi P). Pea3A T and Nos T were used as terminators.