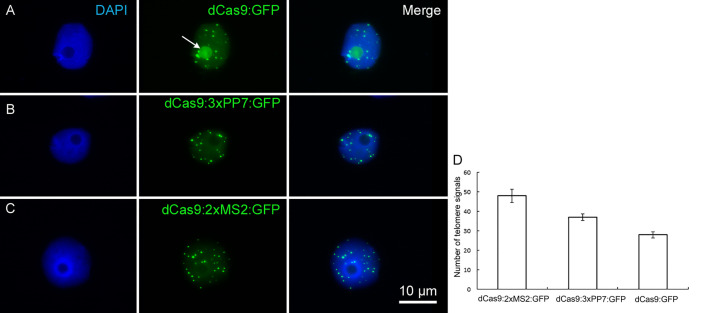
Figure 2.
Live imaging of telomeres in N. benthamiana leaf cells during interphase by CRISPR/dCas9. The distribution of telomeres recognized by (A) dCas9:GFP, (B) dCas9:3xPP7:GFP, and (C) dCas9:2xMS2:GFP. Note, aptamer-based imaging constructs (dCas9:3xPP7:GFP and dCas9:2xMS2:GFP) did not label nucleoli, while the application of dCas9:GFP does (nucleolus shown with white arrow). Nuclei are counterstained with DAPI (1.5 µg/ml) in VECTASHIELD. (D) Diagram showing the efficiency of indirectly and directly labeled dCas9 for targeting telomeric regions. The number of telomere signals was determined based on 20 nuclei per construct. dCas9 indirectly labeled either with MS2 or PP7 aptamers shows more telomeres (p < 0.05).