Abstract
Allele-specific RNA silencing has been shown to be an effective therapeutic treatment in a number of diseases, including neurodegenerative disorders. Studies of allele-specific silencing in hypertrophic cardiomyopathy (HCM) to date have focused on mouse models of disease. We here examine allele-specific silencing in a human-cell model of HCM. We investigate two methods of silencing, short hairpin RNA (shRNA) and antisense oligonucleotide (ASO) silencing, using a human induced pluripotent stem cell-derived cardiomyocyte (hiPSC-CM) model. We used cellular micropatterning devices with traction force microscopy and automated video analysis to examine each strategy’s effects on contractile defects underlying disease. We find that shRNA silencing ameliorates contractile phenotypes of disease, reducing disease-associated increases in cardiomyocyte velocity, force, and power. We find that ASO silencing, while better able to target and knockdown a specific disease-associated allele, showed more modest improvements in contractile phenotypes. These findings are the first exploration of allele-specific silencing in a human HCM model and provide a foundation for further exploration of silencing as a therapeutic treatment for MYH7-mutation-associated cardiomyopathy.
Keywords: allele-specific, hypertrophic cardiomyopathy, RNA silencing
INTRODUCTION
RNA silencing has made recent strides toward clinical application, including antisense oligonucleotide therapeutics used in neurodegenerative disorders (23) and the recent approval of patisiran by the FDA as the first approved RNA interference-based drug (1). These types of therapeutic strategies, which target disease at the RNA level, have the potential to be effective mechanisms of silencing disease-associated molecules before they can confer negative phenotypic effects, rather than treating symptoms after disease progression. One potential area where this type of strategy may be effective is in cardiovascular disease, including cardiomyopathies.
Hypertrophic cardiomyopathy (HCM), characterized by myocardial hypertrophy and disarray, affects 1:500 in the population and can lead to atrial fibrillation, heart failure, and sudden death (24). Causative genetic mutations can be identified in 30–60% of sequenced patients, and 75% of those appear in one of two genes: MYH7 (myosin heavy chain 7) or MYBPC3 (cardiac myosin binding protein c), key components of the cardiac sarcomere (8, 13). The first identified HCM-associated genetic mutation is a single base pair change that causes an arginine to glutamine change in at amino acid 403 (MYH7-R403Q) (14). Molecular analyses of this mutation have over time revealed that it likely causes disease via a dominant-negative “poison-peptide” mechanism, whereby mutated protein products interfere with proper sarcomere function. Single-molecule, dual-beam optical trap studies of human myosin have found that the R403Q mutation causes a decrease in the intrinsic force of the myosin protein (29), while other studies, including examination of the effects of a small-molecule inhibitor of sarcomere power in mice, have indicated that the mutation results in an increase in sarcomere power output and hyperdynamic contraction (15). Recent studies have provided additional insight into the potential mechanisms of MYH7 based HCM, including how mutations in MYH7 may cause hypercontractility of the sarcomere by increasing the number of myosin heads available to interact with actin (3, 4, 36).
Allele-specific RNA silencing has shown therapeutic promise in scenarios where total gene knockdown may be detrimental. In these situations, targeted silencing of a disease-associated allele has shown potential to relieve disease phenotypes, including in some neurodegenerative diseases (37, 40). Allele-specific silencing has also shown promising results in mouse models of cardiovascular disease, including catecholaminergic polymorphic ventricular tachycardia (6) and HCM (17, 48). In humans, the dominant myosin isoform in the adult ventricle is β-myosin heavy chain, encoded by MYH7. Yet in the adult mouse, the dominant isoform is α-myosin heavy chain, encoded by MYH6. Murine studies of HCM have therefore focused on silencing and modulation of MYH6 expression (17). While informative, results of these studies can be hard to translate to potential human therapeutics, as α- and β-myosin have different contractile properties, and silencing of one does not necessarily recapitulate the phenotypic effects of silencing the other. For example, the same R403Q mutation in murine MYH6 and MYH7 showed different functional contractile consequences in in vitro protein motility and enzymatic assays (22). Though the R403Q mutation is one of the most studied mutations in HCM, the vast array of literature focused on the protein-level effects of this mutation has yet to agree on its effects on myosin function, likely due to the issues surrounding species, tissue, and isoform-level differences in myosin function, as well summarized by Nag et al. (29). The genetic background in which these mutations are studied is therefore critical to appropriate interpretation of potential therapeutic strategies, and translational work must therefore aim to study the mutation in a model as close to the appropriate human genetic background as possible. One potential model for such studies is the use of patient-derived human induced pluripotent stem cells (hiPSCs).
Prior work in long QT syndrome has shown that allele-specific silencing of disease-associated mutations can rescue disease phenotypes in human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) (25). hiPSCs have become a widely used system for modeling a number of diseases in vitro (19). Their derivation to cardiomyocytes to study cardiovascular diseases, including hypertrophic cardiomyopathy, has allowed the field to study molecular and cellular phenotypes of disease and test potential therapeutic interventions. This allows for investigation in a human cell context with an unlimited source of cells, rather than working in humanized animal models or with limited human biopsy tissue (11, 18, 20, 21, 28, 41).
To appropriately study patient-derived hiPSC-CMs, assays have been developed to study multiple hypertrophy-associated phenotypes, including contractile defects. Traction force microscopy, which measures the forces cells apply to a substrate by measuring the displacement of substrate embedded fiducials, has become a robust tool for quantifying force, velocity, and power in hiPSC-CMs, leading to discoveries about hiPSC-CM development, maturity, and response to environmental triggers (32, 34, 46). More recent techniques have involved studying the contraction of cells using bright-field videos, removing the need for more extensive microscopic setups and allowing for the analysis of both individual cells and larger tissue-like structures (16). These methods of studying contractility in our cell model system allow us to interrogate the effects of silencing at a single-cell level and gain more accurate insights into how novel therapeutics may ameliorate key disease phenotypes.
Here, we derive hiPSCs from siblings with severe HCM caused by the heterozygous MYH7-R403Q familial mutation to create a human in vitro model of disease. We examine two potential methods of allele-specific gene silencing for both their ability to specifically silence a disease-causing allele in this model and for their ability to relieve disease phenotypes. This study is the first exploration of allele-specific silencing in a human model of HCM.
MATERIALS AND METHODS
MYH7-R403Q-induced pluripotent stem cell derivation and culture.
hiPSCs were derived from two siblings with a heterozygous MYH7-R403Q mutation. hiPSCs for the first line were derived from patient fibroblasts collected under Stanford Institutional Review Board GAP (Genetics and Proteomics) approval number 4237 and cultured under Stanford SCRO (Stem Cell Research Oversight) 568 (Fig. 1A, patient 1). Additional experiments were carried out under Stanford Protocol 2663. Derivation of patient-specific hiPSC lines was performed as previously described (20, 45). Fibroblasts were passaged before infection with lentiviral vectors containing the four Yamanaka factors (OCT4, SOX2, KLF4, and c-MYC). hiPSC colonies were maintained on Matrigel-coated plates (BD Biosciences) in mTESR-1 medium (StemCell Technologies) (20). Immunofluorescent staining showed expression of the pluripotency markers in the created hiPSCs (Supplementary Fig. S1, https://doi.org/10.6084/m9.figshare.11704254). These cells were used for all silencing experiments except Fig. 2, C–G. Experiments in Fig. 2, C–G, were conducted on a second line from a sibling (Fig. 1A, patient 2) who also carried the MYH7-R403Q mutation. hiPSCs and hiPSC-CMs for this line were derived by Cellular Dynamics International (Line 01178.103). hiPSCs were maintained on 1:200 Matrigel plates in mTesr. The wild type control cell line used for all ASO silencing experiments was line SCVI273 obtained from the Stanford Cardiovascular Institute Biobank.
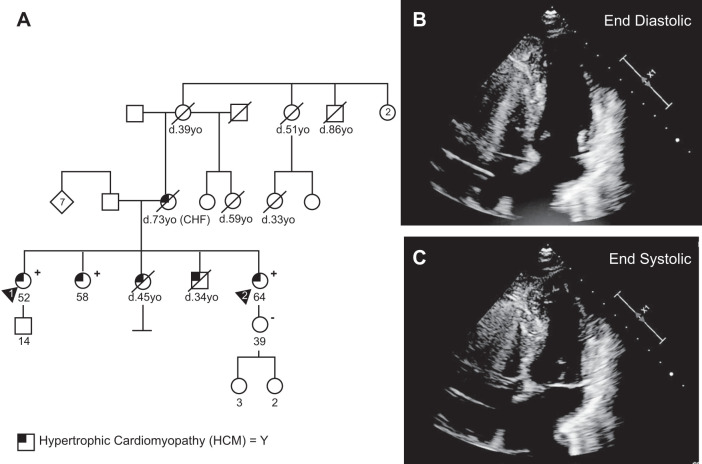
Fig. 1.
Family pedigree and hypertrophic phenotype of MYH7-R403Q hypertrophic cardiomyopathy (HCM) patient. A: family pedigree for patient 1 (designated by arrowhead 1) from whom the MYH7-R403Q human induced pluripotent stem cell (hiPSC) line was created. The patient and all four of her siblings showed symptoms of HCM. Two living siblings also tested positive for the R403Q mutation. One of the siblings (patient 2, designated by arrowhead 2) additionally donated cells for hiPSC derivation, which were used in the experiments in Fig. 2, C–G. B, C: apical four-chamber echocardiography images of patient 1. Images demonstrate small cavity, left ventricular hypertrophy, and abnormal coaptation of the mitral valve, as well as systolic anterior motion of the mitral valve. Extraneous computer graphics and heart-rate information were removed from echocardiography images.
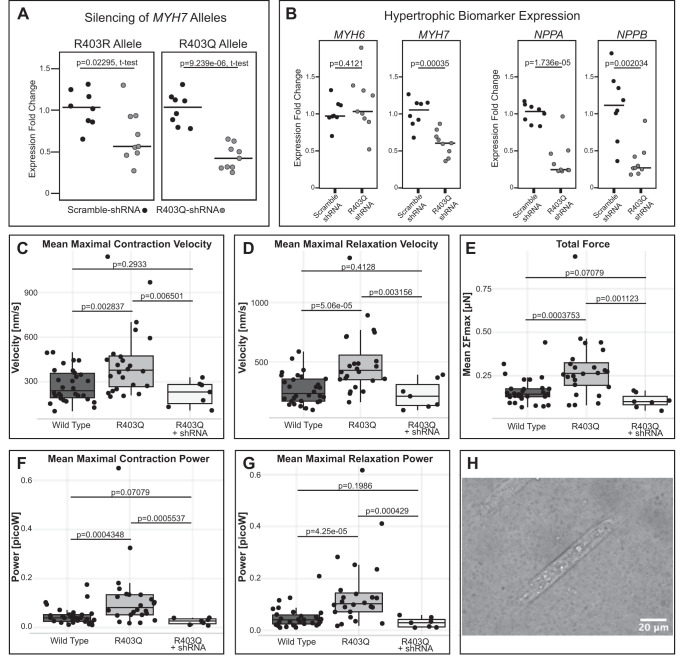
Fig. 2.
shRNA knockdown reduces hypertrophic biomarker expression and contractile disease phenotypes in MYH7-R403Q human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs). A: heterozygous MYH7-R403Q hiPSC-CMs transduced with a virally delivered R403Q-targeting shRNA (H10.8L) show reduced expression of both the R403Q and R403R alleles as compared with hiPSC-CMs transduced with a scrambled shRNA. Individual data points and medians are plotted, expression normalized to scramble-treated RNA expression for either the R403R or R403Q allele individually. hiPSC-CMs were shRNA-treated on day 20 of differentiation, and RNA was collected after 5 days of treatment. B: MYH7-R403Q hiPSC-CMs treated with this same R403Q-targeting shRNA show reduction of overall MYH7 RNA levels, but no change in MYH6 expression levels, as well as a decrease in RNA expression of hypertrophic biomarkers NPPB and NPPA. Individual data points and medians are plotted, expression normalized to scramble-treated RNA expression. All P values are from a two-sided t test. C–G: untreated MYH7-R403Q hiPSC-CMs from a sibling cell line (CDI Line 01178.103) show increased velocity, force, and power during contraction and relaxation compared with wild-type (WT) hiPSC-CMs, as measured by traction force microscopy. Viral transduction with the R403Q-targeting H10.8L shRNA reduces these contractile measures back to wild-type levels. H: example image of a WT hiPSC-CM on a cellular micropatterning device. These Matrigel patterns were printed onto a polyacrylamide gel with embedded fluorescent beads to track contractile motion.
hiPSC-cardiomyocyte differentiation.
hiPSCs were differentiated to cardiomyocytes using a modified version of Sharma et al. 2015 (39). Briefly, hiPSCs were cultured until they reached 80% confluence in 12-well dishes. For ASO phenotyping experiments: hiPSCs were then exposed to differentiation media (RPMI with B27-insulin) for 2 days with the addition of GSK3 inhibitor CHIR. Optimal CHIR concentrations varied between cell lines. Each new cell line was tested with concentrations ranging from 3 to 8 μM. Cells were then exposed to differentiation media with the addition of Wnt inhibitor IWR for 2 days. After that, cells were cultured in the media alone for 2 days before changing to RPMI with complete B27 for 3 days. hiPSC-CMs were then maintained in starvation media (RPMI minus glucose with B27 and lactate added). This helped to purify and mature only the hiPSC-CMs, which can metabolize lactate in the absence of glucose. hiPSC-CMs were cryopreserved in BamBanker and stored in liquid nitrogen. For shRNA experiments and Fig. 3A, differentiation differed slightly with a single day “rest” period between CHIR and IWR exposure, and with maintenance of hiPSC-CMs in media containing glucose after day 20.
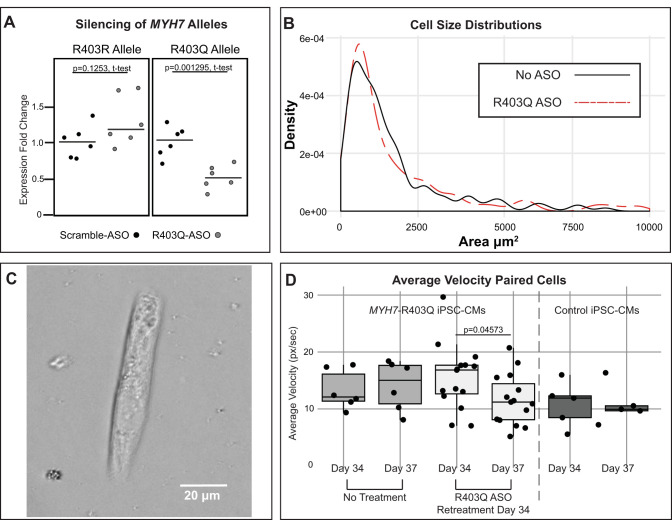
Fig. 3.
R403Q-Targeting antisense oligonucleotide (ASO) reduces contractile velocity but not cell size in MYH7-R403Q induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs). A: heterozygous MYH7-R403Q human (h)iPSC-CMs transfected with an R403Q-targeting ASO show decreased expression of the R403Q allele (49% reduction, P = 0.001295) but not the R403R allele. Individual data points and medians are plotted, expression normalized to scramble-treated RNA expression for either the R403R or R403Q allele individually. P values are from a two-sided t test. hiPSC-CMs were at differentiation day 21 at transfection, treated with 2 μM ASO, and collected for RNA extraction after 24 h. The R403Q-targeting ASO sequence is mUmCmACCTGAGGmGmU, where mN indicates 2′-O-methylated RNA bases and DNA base core is in boldface. The ASO additionally has a phosphorothioate backbone. The underlined T represents the single base that discriminates the R403Q allele from the R403R allele. The scrambled control ASO used in this experiment is a 20 bp gap-mer with a fluorescent label: /56-FAM/mUmGmAmCmGTTGTACGACGmCmAmUmUmC. B: cell size density plots from the MYH7-R403Q cell line treated with transfection-reagent only, and the MYH7-R403Q cell line treated with the R403Q-targeting ASO during differentiation. hiPSC-CMs treated with the R403Q-targeting ASO showed a slight decrease in the median population cell size as compared with hiPSC-CMs treated with the transfection reagent alone [R403Q-ASO median = 916 μm2, n = 146 cells vs. cells treated with only transfection reagent (No ASO) median = 1,077 μm2, n = 144 cells]. Plotting bandwidth adjustment = 0.8. C: example image of an ASO-treated MYH7-R403Q hiPSC-CM on cellular micropatterning device used to analyze cellular motion in D. These Matrigel patterns were printed onto glass-bottom dishes. The cell shown in this image is from differentiation day 34. D: MYH7-R403Q hiPSC-CMs were treated with the R403Q-targeting ASO at days 6, 12, and 18 of differentiation and plated onto protein micropatterns on glass-bottom dishes on day 29. Videos taken 5 days later at day 34 showed that the ASOs did not have lasting effects on contraction as compared with hiPSC-CMs that received no treatment. hiPSC-CMs were retreated with the R403Q-targeting ASO after measurements were taken on day 34, and videos were taken again 3 days later. Plotted are paired hiPSC-CMs, where we could confidently identify the same beating cell in videos taken before and after treatment. Each point represents the average contractile velocity of a single cell over a 15 s video measured in pixels per second. Untreated MYH7-R403Q hiPSC-CMs show a slight increase in contractile velocity over this 3-day time period, while our ASO-retreated cell population shows a decrease in contractile velocity. Age-matched wild-type control hiPSC-CMs show little change in contractile velocity over this 3-day time period.
hiPSC-cardiomyocyte shRNA silencing experiments.
hiPSC-CMs were seeded into a 24-well plate at a density of 5 × 105 cells per well. hiPSC-CMs were transduced at differentiation day 20 in 300 μl of RPMI/B27 media using 3.9e9vg of adeno-associated virus (AAV)6-scramble-shRNA or AAV6-H10.8L shRNA expressing virus (Supplementary Fig. S2, https://doi.org/10.6084/m9.figshare.11704278). Nine wells were transfected with the scramble-shRNA virus, while 10 wells were transfected with the H10.8L virus. hiPSC-CMs were collected after 5 days, and RNA was extracted with the Qiagen miRNeasy kit, with additional DNase treatment step. cDNA was made with the Applied Biosystems High Capacity cDNA kit. Quantitative (q)PCR was performed as described below. One sample from each condition in Fig. 1, A and B, was excluded due to high outlier values in MYH7 qPCR and abnormal housekeeping gene qPCR values. The H10.8L-containing AAV6 virus was produced at Stanford’s Neuroscience Gene Vector and Virus Core, while the scramble-shRNA-containing AAV6 virus was produced by Virovek (Hayward, CA).
qPCR analysis of MYH7 alleles.
Allele-specific qPCR was performed using mutant or wild type-specific forward primers and a common reverse primer, as previously described in Wheeler et al. (45). Each forward primer contains a mismatch at the penultimate nucleotide to increase allele specificity. The MYH7-specific fluorescent probe was optimized for maximum sequence dissimilarity from MYH6. Allele-specific qPCR conditions using TaqMan Fast Universal PCR Master Mix: 95°C 20”, 40 cycles of 95°C 30”, 58°C 20”, 72°C 30” for R403Q or 40 cycles of 95°C 30”, 64°C 20”, 72°C 30” for R403R. Different annealing temperatures were used to increase the allele-specificity of our forward primers. Due to the differing reaction conditions, expression was not directly compared between the two alleles. Instead, allele expression was individually normalized to scramble-treated cells using the delta-delta Ct method. Endogenous control was EEF1A2. R403R Forward Primer: 5′ GGGCTGTGCCACCCTAA 3′, R403Q forward primer: 5′GGGCTGTGCCACCCTAG 3′, common reverse primer: 5′CGCGTCACCATCCAGTTGAAC 3′; MYH7-specific fluorescent probe: FAM-5′TGCCACTGGGGCACTGGCCAAGGCAGTG 3′-TAMRA.
RNA extraction and cDNA amplification.
For some early experiments, RNA was extracted using the Qiagen miRNeasy Kit with on-column DNase treatment. However, for the majority of experiments, RNA was extracted using TRIzol-chloroform and treated with the Thermo Fisher Turbo DNase kit. cDNA amplification was achieved using the High Capacity cDNA Reverse Transcription Kit with RNase Inhibitor. For some reactions, half of the cDNA was digested with AvaI (to specifically cut the wild-type allele), and the other half was cut with Bsu36I (to specifically cut the mutant allele) before their individual qPCR reactions, though we later determined this step to be nonessential.
qPCR of hypertrophic genes and biomarkers.
qPCR of common hypertrophic and biomarker genes was performed using IDT predesigned probes. Probes used were: EEF1A2 (Hs.PT.58.3514123), MYH6 (Hs.PT.58.2106207), MYH7 (Hs.PT.58.14589334), NNPA (Hs.PT.58.4259173), and NPPB (HS.PT.58.19450190). qPCR was performed using IDT’s PrimeTime Master Mix using its recommended protocol on a ViiA 7 Real-Time PCR System (Thermo Fisher Scientific). Data were analyzed using the delta-delta Ct method.
Micropatterning of single cells on polyacrylamide gel surfaces and analysis of contractility from traction force microscopy.
Micropatterning extracellular proteins on a surface for cell culture can set the morphology of single cells to match the same shape of the micropatterned region. Culturing single hiPSC-CMs on micropatterns with a rectangular shape forces these cells to have a more mature morphology and organization of the sarcomere-based contractile machinery (32). In addition, micropatterning hiPSC-cardiomyocytes with a rectangular shape on a soft material with physiological tissue rigidity further enhances the physiology of their contractile function and allows measurement of contractility by tracking the cell-induced deformation of the material under each cell (32, 34). MYH7-R403Q hiPSC-cardiomyocytes from CDI Line 01178.103 and healthy control line 01027.101 were cultured on 2,000 μm2 rectangular (7:1 aspect ratio) Matrigel patterns on polyacrylamide (PA) surfaces as previously described in Ribeiro et al. (7, 32) and (45). Soft lithography was used to fabricate PDMS microstamps (31). Microstamps were flooded with 1:10 dilution of Matrigel in cold L15 medium at 4°C for 24 h and dried using N2 after aspirating and washing the surface twice with cold L15 medium. Patterns were stamped onto a plasma-treated glass coverslip. Gels were fabricated as reported elsewhere (12, 30, 45). PA gel components (12% acrylamide, 0.15% N, N-methylene-bis-acrylamide) and fluorescent microbeads were mixed in DI water; 50 µl of the solution was added to clean, pretreated coverslips. Ammonium persulfate was used as a catalyst for gel polymerization, and TEMED was used as an initiator. Micropatterned coverslips were then placed on top of the gel solution. After polymerization, coverslips were removed to reveal the patterns, and hiPSC-CMs were seeded onto them in appropriate culture media. MYH7-R403Q hiPSC-CMs were transduced with 3.9e10vg of AAV6-H10.8L-shRNA or left untreated. Videos of beating hiPSC-CMs were acquired with a Zeiss Axiovert 200M inverted microscope equipped with a Zeiss Axiocam MRm CCD camera to image the displacement of fluorescent microbeads in polyacrylamide materials supporting the adhesion of micropatterned cells. The microscope had an environmental chamber (PeCon) to keep temperature of cell media at 37°C. Videos of beating hiPSC-CMs were acquired while electrically pacing the hiPSC-CMs with 10 ms-wide bipolar pulses of electric-field stimulation at 10–15 V with a frequency of 1 or 2 Hz (Myopacer, IonOptix). Only cells beating at the appropriate frequency were analyzed. We measured the parameters of cell contractility from traction force microscopy results following the methodology reported by Ribeiro et al. (33).
hiPSC-CM staining for cell-size measurements.
hiPSC-CMs were stained for α-actinin (to confirm cardiomyocyte identity), actin, and DAPI for cell-size measurements. Briefly, beating hiPSC-CMs plated on Matrigel-coated coverslips were relaxed in 50 mM KCl before fixation with 4% paraformaldehyde. hiPSC-CMs were permeabilized with 0.1% Triton X-100 and incubated in 5% normal goat serum before overnight incubation with the primary α-actinin antibody (Sigma Aldrich, A7811) at 4°C. After being washed with 0.1% Tween 20, hiPSC-CMs were incubated for 1 h with the secondary antibody (Alexa Fluor 568, Sigma Aldrich, A11031). After being washed again with Tween 20, hiPSC-CMs were incubated in ActinGreen for 10 min before being mounted with Prolong Diamond Antifade Mountant with DAPI. The next day, hiPSC-CMs were imaged using a Nikon Eclipse 90i microscope. Cell-size measurements were calculated by blinded circling of hiPSC-CMs in ImageJ. Cell sizes greater than 10,000 μm2 were removed before analysis.
ASO silencing in hiPSC-CMs.
ASO silencing experiments used different ASOs, concentrations, and transfection reagents as described in the text. However, the base protocol is as follows. ASOs were custom designed and ordered from IDT (Integrated DNA Technologies, Coralville, IA) before resuspension to 100 μM in sterile H2O. hiPSC-CMs or differentiating hiPSCs were plated in 12- or 24-well tissue culture plates on 1:200 Matrigel. After 2–3 days, culture medium was removed and replaced with fresh medium. For each ASO, a master mix was prepared. The transfection reagent (TransIT-TKO, Mirus Bio) was added to fresh Opti-MEM and mixed gently. The ASO was added to the desired concentration and mixed gently by pipetting before sitting at room temperature for 30 min. As an example, for 1 well of 6 μM ASO in a 12-well plate, 49 μl of Opti-MEM was mixed with 3 μl of TransIT-TKO before the addition of 48 μl of ASO. After 30 min, 100 μl of the master mix was added dropwise to the appropriate well of the tissue culture dish, which contained 700 μl of fresh media. The culture plate was gently shaken to disperse the ASO complexes and returned to 37°C for at least 24 h. When collected for RNA analysis, hiPSC-CMs were either dissociated with accutase, rinsed in PBS, and centrifuged or collected with TRIzol. Pellets or TRIzol collections were snap-frozen in liquid nitrogen before RNA extraction. ASOs were ordered from Integrated DNA Technologies and have a phosphorothioate backbone. R403Q-targeting ASO sequence: mUmCmACCTGAGGmGmU where mN indicates 2′-O-methylated RNA bases and DNA base core is in boldface. Scramble ASO sequence used in Fig. 3A, designed using InvivoGen’s siRNA wizard: /56-FAM/mUmGmAmCmGTTGTACGACGmCmAmUmUmC.
Silencing through differentiation treatments.
ASO silencing treatments during differentiation were performed as described above, except that treatments were delivered throughout differentiation at days 6, 12, and 18 in 12-well dishes. The day 6 treatment consisted of 6 μM ASO and 3 μl of TransIT-TKO, while the day 12 and day 18 treatments were halved (3 μM ASO and 1.5 μl TransIT-TKO per well) to avoid toxicity due to repeated high exposure to transfection reagent.
Cellular micropatterning on glass for velocity analysis.
Microcontact printing of rectangular protein patterns was performed as previously described (2). Briefly, PDMS microstamps containing 17 × 119 mm2 rectangles were used to microprint dilute Matrigel onto glass bottom six-well plates. hiPSC-CMs were replated onto micropatterns at 10,000–50,000 cells/mL. Videos of patterned hiPSC-CMs were acquired using a Keyence BZ-X microscope. Videos were taken using a ×40 objective for 15 s at 29 frames per second. Videos were analyzed as described in Huebsch et al. (16). Average maximum velocity was calculated for each cell. For each cell with an average maximum velocity above 30 pixels/second, a secondary round of examination was performed to remove any cell suspected of being two hiPSC-CMs attached to the same pattern. These doublet hiPSC-CMs were removed from all further analysis.
Statistical analysis.
All statistical analysis was performed using RStudio. qPCR gene and allele expression data was analyzed using Student’s t test. Contractility measurements in Fig. 2, C–G, were analyzed using a two-tailed Mann-Whitney U test. Cell size measurements in Fig. 3B were analyzed using a Wilcoxon rank-sum test without continuity correction. Velocity measurements in Fig. 3D were analyzed using Student’s two-sided t test.
RESULTS
hiPSC derivation from a family with multiple affected HCM patients.
We derived hiPSCs and differentiated hiPSC-CMs from two members of a family with HCM. A family of five siblings with HCM showed signs of severe disease. Two of the five siblings passed away at young ages (34, 45). The three remaining siblings all tested positive for the MYH7-R403Q mutation. One sibling suffered end-stage heart failure and received a heart transplant at the age of 41 yr. A more extensive family history revealed that the mother of the five siblings also had HCM and died at the age of 73 yr. While she did not undergo genetic testing, a number of additional early deaths in the maternal lineage (at the ages of 33, 39, 51, and 59 yr) suggest penetrance across multiple generations. Our primary cell line was derived from a 45 yr old female proband with the disease-associated MYH7-R403Q mutation (Fig. 1A, patient 1, 52 yr old at time of publication). The patient was diagnosed with HCM after presenting with a heart murmur at the age of 13 yr. A second hiPSC cell line was derived by Cellular Dynamics from an affected sibling (Fig. 1A, patient 2).
In vitro shRNA silencing of R403Q allele in human hiPSC-CMs achieves knockdown and decrease in hypertrophic biomarkers.
We used virally delivered short hairpin RNA (shRNAs) to silence a dominant, HCM-associated mutation, MYH7-R403Q, in patient-derived hiPSC-CMs to show mutant allele knockdown in human cells. Potential shRNA designs were tested in a fluorescent-reporter HEK cell model to arrive at the mutant-allele-specific H10.8L shRNA design (Supplementary Fig. S3, https://doi.org/10.6084/m9.figshare.11704362). MYH7-R403Q hiPSC-CMs were transduced with an AAV6 viral vector expressing the H10.8L shRNA under the control of an H1 promoter (Supplementary Fig. S4, https://doi.org/10.6084/m9.figshare.11704737). Five days posttransduction, hiPSC-CMs showed a significant decrease in expression of the mutant allele RNA by allele-specific qPCR as compared with hiPSC-CMs transduced with an AAV6 vector expressing a scrambled control shRNA (Fig. 2A, 57% average knockdown of mutant allele, P = 9.239e-06) as well as significant but lesser knockdown of the wild-type allele (Fig. 2A, 33% average knockdown of the wild-type allele, P = 0.02295). These hiPSC-CMs also showed a significant decrease in their overall MYH7 levels (Fig. 2B, P = 0.00035) while maintaining MYH6 levels comparable to untreated hiPSC-CMs (Fig. 2B, P = 0.4121). They additionally showed significant decreases in RNA expression of hypertrophic biomarkers NPPA and NPPB (Fig. 2B, P = 1.736e-05 and 0.002034, respectively).
In vitro shRNA silencing in R403Q hiPSC-CMs shows amelioration of contractile phenotypes.
MYH7-R403Q hiPSC-CMs from a sibling cell line derived by Cellular Dynamics (CDI Line 01178.103) were cultured on protein micropatterning devices and analyzed via high-speed imagery for functional profiling using traction force microscopy. Analyses revealed that when compared with wild-type hiPSC-CMs, these MYH7-R403Q hiPSC-CMs had significantly increased maximal contraction and relaxation velocities and powers, as well as significantly increased maximal force (Fig. 2, C–G). Populations treated with the AAV6-H10.8L shRNA showed significant decreases back to wild-type levels across all measured parameters, showing an amelioration of disease phenotype.
In vitro ASO silencing in R403Q hiPSC-CMs shows specific knockdown of the R403Q allele.
We designed allele-specific MYH7-R403Q-targeting antisense oligonucleotides (ASOs) based on design parameters from prior publications (9, 42). Our final ASO design comprised a 12-mer composed of 2′ O-Methyl modified RNA wings for stability, a 7-base DNA core, and a phosphorothioate backbone. When transfected into the MYH7-R403Q hiPSC-CMs, we saw a 49% decrease in expression of the R403Q allele (Fig. 3A, P = 0.001295), while we saw no significant decrease in the wild-type R403R allele as compared with hiPSC-CMs treated with a scrambled ASO. This decrease in expression was also examined using Sanger sequencing, where PCR on equal concentrations of cDNA revealed a decrease in overall expression of this region at 24 and 48 h after sequencing with the R403Q-targeting ASO, as well as a decrease in the Sanger trace corresponding with the R403Q allele, indicating that this allele was being silenced to a greater degree than the R403R allele (Supplementary Fig. S7, https://doi.org/10.6084/m9.figshare.11704797). We also performed the same experiment examining a heterozygous SNP present on the R403Q allele and saw the same decrease in R403Q allele expression (Supplementary Fig. S7, https://doi.org/10.6084/m9.figshare.11704797).
Allele-specific ASO silencing during differentiation shows modest effects on hypertrophic cell size.
Myocardial overgrowth in HCM hearts is characterized by an increase in individual cell size rather than an increase in the number of cardiomyocytes present. We aimed to reduce pathologic cell size in our MYH7-R403Q hiPSC-CMs via treatment with R403Q-targeting ASOs. Untreated MYH7-R403Q hiPSC-CMs fixed and stained at day 30 showed an overall increase in cell population size as compared with wild-type hiPSC-CMs (Supplementary Fig. S8, https://doi.org/10.6084/m9.figshare.11704806, MYH7-R403Q hiPSC-CM median size = 1,076 μm2, n = 147, vs. SCVI273 control cell line, median size = 808 μm2, n = 150, P = 0.002207, Wilcoxon rank-sum test without continuity correction).
We treated MYH7-R403Q hiPSC-CMs during differentiation, starting with a 6 μM ASO treatment at day 6 of differentiation, and 3 μM ASO treatments at day 12 and day 18. hiPSC-CMs were sparsely replated on day 20 and fixed at day 30 for staining of α-actinin. hiPSC-CMs treated with the R403Q-targeting ASO showed only a modest, nonsignificant decrease in the median population cell size as compared hiPSC-CMs treated with the transfection reagent alone [Fig. 3B, R403Q-ASO median = 916 μm2, n = 146 cells, vs. cells treated with only transfection reagent (no ASO) median = 1077μm2, n = 144 cells].
Allele-specific ASO silencing reduces contractile phenotypes of disease.
To test the effects of the R403Q-targeting ASO on contractile phenotypes of disease, MYH7-R403Q hiPSC-CMs were treated with the silencing ASO during differentiation and plated on Matrigel micropatterns printed on glass culture dishes. Untreated MYH7-R403Q hiPSC-CMs showed an increase in average contractile velocity as compared with wild-type hiPSC-CMs (Supplementary Fig. S9, https://doi.org/10.6084/m9.figshare.11704815). MYH7-R403Q hiPSC-CMs treated during differentiation showed no relief in contractile phenotypes at day 34 of differentiation (Fig. 3D). However, MYH7-R403Q hiPSC-CMs that were retreated with the R403Q-targeting ASO at day 34 and imaged at day 37 showed a decrease in average contraction velocity over the 3-day time period (Fig. 3D). In contrast, untreated MYH7-R403Q hiPSC-CMs and wild-type hiPSC-CMs showed no significant change in velocity over this 3-day period (Fig. 3D). Average contractile velocity of paired hiPSC-CMs (average ± SE): Untreated MYH7-R403Q hiPSC-CMs, day 34 average: 13.3 ± 1.4 px/s, n = 6; untreated MYH7-R403Q hiPSC-CMs, day 37 average: 14.1 ± 1.8 px/s, n = 6; day 34 vs. day 37 P value = 0.7358. R403Q-ASO treated MYH7-R403Q hiPSC-CMs day 34 average: 15.5 ± 1.5 px/s, n = 15. R403Q-ASO retreated MYH7-R403Q hiPSC-CMs; day 37 average: 11.6 ± 1.2 px/s, n = 15; day 34 vs. day 37 P value = 0.04573; wild-type control hiPSC-CMs, day 34 average: 10.8 ± 1.8 px/s, n = 5; wild-type control hiPSC-CMs, day 37 average: 10.7 ± 1.5 px/s, n = 5; day 34 vs. day 37 P value = 0.9667.
DISCUSSION
Current therapies for HCM include invasive procedures such as implantation of a defibrillator, surgical removal of heart muscle tissue (myectomy), and heart transplantation. Genetic therapeutics designed to target disease-associated alleles could present alternative, less invasive strategies of combating disease. Previous investigation of such therapeutics in HCM have focused on silencing genes in a murine model, rather than a human model. However, the dominant myosin isoforms are different between mice and humans. Here, we present the first examination of allele-specific silencing in a human disease model of HCM. We explore two potential genetic therapeutics targeting a heterozygous disease-causing mutation in MYH7 in a human iPSC-CM model of disease, using biophysical assays to assess their impacts on contractile phenotypes of disease.
We derived an hiPSC line from a patient with familial HCM to create our disease model. Previous studies using patient-derived hiPSC-CMs have shown them to be a useful model for studying disease phenotypes in the laboratory and have proven to be an important platform for studying potential silencing therapeutics in cardiovascular disease (20, 25). The patient from whom our cell line was derived was diagnosed with nonobstructive HCM at age 13 yr and was recommended to have an implantable cardioverter defibrillator placed at age 39 yr because of nonsustained ventricular tachycardia. Genetic testing revealed a heterozygous R403Q mutation in MYH7. This patient additionally had a strong family history of early death due to heart failure from HCM, as well as family members who additionally carried the R403Q mutation and one who had undergone a heart transplant.
To assess allele-specific silencing via shRNA of an HCM-associated gene in this model, we virally introduced an shRNA designed to specifically target the MYH7-R403Q allele. After viral transduction with the R403Q-targeting shRNA, hiPSC-CMs containing the heterozygous MYH7-R403Q mutation showed 57% knockdown of the mutant allele as measured by allele-specific qPCR. We additionally saw a decrease in molecular markers of hypertrophy, including NPPA and NPPB. Previous studies of allele-specific MYH6 silencing in mice have shown similar decreases in NPPA and NPPB expression, further supporting the idea that reducing mutant myosin transcripts may have beneficial effects on other cardiomyopathy-related gene expression (17). While we do see a 41% reduction of overall MYH7, we do not see a reduction in MYH6, indicating that our shRNA treatment is specific to MYH7 and does not cause off-target silencing of MYH6.
To assess functional improvement in phenotype of shRNA-treated hiPSC-CMs, we assayed contractile function of a related MYH7-R403Q hiPSC line. We deployed cellular micropatterning devices on hydrogel substrates to measure contractile properties of hiPSC-CMs using traction force microscopy [as described in Ribeiro et al. (32)]. Previous studies of hiPSC-CMs and cardiac tissues derived from patients with HCM have shown contractile dysfunction, including contractile arrhythmias and hypercontractility (5, 20, 38), as well as greater developed force and shorter twitch duration (10), in patient-derived cardiomyocytes as compared with wild-type controls. In our study, we found that MYH7-R403Q hiPSC-CMs had significantly increased maximal contraction and relaxation velocities and powers, as well as significantly increased maximal force when compared with wild-type hiPSC-CMs. We saw a shift toward wild-type hiPSC-CM levels of all measured parameters in MYH7-R403Q hiPSC-CMs after shRNA treatment, as compared with nontransfected MYH7-R403Q hiPSC-CMs. While our shRNAs reduced expression of not only the mutant (R403Q) but also the wild-type (R403R) MYH7 allele in our in vitro studies, this overall drop in MYH7 did not prevent the return to normal phenotype in our silenced hiPSC-CMs, leading us to believe that the small drop in MYH7-R403R RNA is tolerated.
We continued to pursue an allele-specific silencing molecule and turned to antisense oligonucleotides, which have shown great promise in both neurodegenerative disorders and cardiovascular disease. We tested a number of gap-mer ASO designs, from 12 to 20 bp in length, with the target variant base either centered or slightly off-center and with or without additional mismatches, before settling on our final 12 bp design, which specifically silenced the R403Q allele (Supplementary Table S1, https://doi.org/10.6084/m9.figshare.11704731, Supplementary Figs. S5, https://doi.org/10.6084/m9.figshare.11704761, and S6, https://doi.org/10.6084/m9.figshare.11704785). Our best-designed ASO showed a 49% decrease in MYH7-R403Q expression with no significant knockdown of the wild-type allele (Fig. 3A). However, while this short ASO design lent itself to the greatest specificity, it does pose potential problems when considering off-target silencing. A recent paper by Yoshida et al. (47) estimated that 12 bp ASOs may have up to 3.4 potential off-target binding regions with zero-sequence mismatches and up to 122 potential off-target binding regions if a single mismatched-base is tolerated. While these numbers are large, they do not take into account potential secondary structure of the RNA, which is known to impact target affinity in ASOs (35), or whether or not the transcripts containing those potential off-target sequences are expressed in the target cell type. Further investigation of the effects of these ASOs on off-target gene expression via whole transcriptome RNA-expression analysis at both the cell and tissue levels would be necessary before this oligonucleotide could be introduced into a clinical setting.
Despite these considerations, we aimed to reduce pathologic cell size in our MYH7-R403Q hiPSC-CMs via treatment with the R403Q-targeting ASOs. We showed that day 30 MYH7-R403Q hiPSC-CMs showed an increase in overall population cell size as compared with a wild-type line (Supplementary Fig. S8, https://doi.org/10.6084/m9.figshare.11704806). We found that treatment of MYH7-R403Q hiPSC-CMs during differentiation with the R403Q-targeting ASO could induce a modest reduction in overall population cell size at day 30 (Fig. 3B), as compared with hiPSC-CMs treated with only the transfection reagent. Previous examinations of cell size in hypertrophic human hearts have shown mean cell diameter changes of around 25% in the left ventricular walls as compared with control hearts (44). Our untreated MYH7-R403Q hiPSC-CMs show a 33% increase in median cell size as compared with the wild-type control line. However, our R403Q-ASO treated hiPSC-CMs show only a 13% increase in median cell size (Fig. 3B), indicating modest relief of phenotype back toward wild-type cell sizes.
As with our shRNA-treated hiPSC-CMs, we aimed to examine the effects of our antisense oligonucleotide silencing on contractile phenotypes in our hiPSC-CMs. We used automated video analysis to assess the impact of the R403Q-targeting ASO on contractile phenotypes of disease. While ASO treatment during differentiation did not have lasting effects on contraction, this was not especially surprising, given that we see persistence of ASO silencing in our hiPSC-CMs for around 1 wk, and the hiPSC-CMs had not received a dose of ASO for 16 days by the analysis time point. However, when we retreated hiPSC-CMs at this time point and imaged again 3 days later, we saw a decrease in the average contractile velocity of hiPSC-CMs treated with our allele-specific ASO (Fig. 3D). We did not see a decrease in the velocity of untreated MYH7-R403Q hiPSC-CMs over the same 3-day period (Fig. 3D). While we examined paired hiPSC-CMs in Fig. 3D, where we were able to confidently match videos of hiPSC-CMs taken at day 34 and day 37, we also present population level data of all beating hiPSC-CMs recorded for these populations in Supplementary Fig. S9, https://doi.org/10.6084/m9.figshare.11704815. This may suggest that ASO silencing is able to relieve contractile phenotypes after cell growth and maturation.
A growing number of investigations of HCM-associated MYH7 mutations have also studied the allele-specific expression of both wild-type and mutant MYH7 alleles and have reported imbalanced expression not only between patients, but between individual cells from the same patient (26, 27, 43). These imbalances have been associated with variable contractile and calcium-handling phenotypes between mutation-containing cardiomyocytes (26), with some cells showing properties quite similar to wild-type cells and others showing highly disordered phenotypes. Montag et al. (26) suggest that these imbalances may be due to cells with high wild-type-allele expression mimicking healthy cells and cells with high mutant-allele expression exhibiting more detrimental phenotypes. This underlying imbalance in expression between cells may contribute not only to the increased difficulty in phenotyping large cell populations, where variance between cells may make population level conclusions noisy, but also to the disconnect between allele specificity and phenotyping effect that we observe in our silenced hiPSC-CM populations.
While the R403Q-targeting shRNA knocked down both alleles of MYH7, it conferred significant benefits to both molecular and contractile phenotypes of disease. Yet while the R403Q-targeting ASO showed more specific knockdown, it also showed more modest phenotypic benefits. This disparity in expected phenotypic response may be due to the dose and delivery of each molecule. AAV viral delivery results in episomal stability of the delivered genetic material, allowing high levels of the shRNA to be transcribed in the cell under the control of the H1 promoter. Conversely, despite delivering our ASOs at high concentrations (6 μM) we found their silencing effects to last for only about 1 wk (Supplementary Fig. S7 URL, https://doi.org/10.6084/m9.figshare.11704797). It may be that persistent treatment with an MYH7 RNA-targeting molecule is needed to sufficiently silence the disease-associated transcripts. This would pose important considerations for future clinical applications, where repeated treatments can become expensive and burdensome to patients and caretakers. Additionally, our R403Q-targeting ASO decreased mutant allele expression by only about half, leaving dominant-negative transcripts present in the cell. While we created a number of other ASOs during the design process that knocked down overall MYH7 to much greater levels, none of them proved to be allele-specific (Supplementary Figs. S5, https://doi.org/10.6084/m9.figshare.11704761, and S6, https://doi.org/10.6084/m9.figshare.11704785). This tradeoff between specificity and overall silencing may be important to consider in situations where even a small amount of mutant protein may be enough to cause disease. Single, efficient treatment with an shRNA-based strategy, as has been done in related animal models of hypertrophic cardiomyopathy (48), may prove to be the most efficacious path forward.
In conclusion, therapeutic gene silencing has the potential to ameliorate disease phenotypes by targeting the underlying genetic cause of disease. Here, we examined two methods of gene silencing in a human iPSC-CM disease model of HCM, shRNA and ASO silencing. We used traction force microscopy and automated video analysis to interrogate the effects of these silencing methods on contractile disease phenotypes. We found dissociation of allelic specificity and functional improvements, suggesting a more complex allelic control underlying the role of MYH7-R403Q in disease expressivity. We demonstrated that while less allele specific, shRNA silencing can significantly improve contractile and molecular phenotypes of disease, potentially due to high, continuous levels of treatment conferred by AAV viral delivery. We additionally showed that while ASO silencing has the potential to improve allele-specificity, it showed more mild relief of phenotypes in our hiPSC-CMs. These findings demonstrate that decreasing expression of MYH7 in a human model of disease has the potential to relieve HCM phenotypes, but that further study may be needed to parse the relationship between therapeutic silencing and mutant and wild-type expression underlying disease phenotypes. We hope that this first exploration of allele-specific silencing in a human HCM model will lay the foundation for continued testing of silencing as a therapeutic treatment for cardiomyopathy.
GRANTS
A. Dainis received support from the National Science Foundation Graduate Research Fellowship Program. E. Ashley received funding from National Institutes of Health (NIH) Director’s New Innovator Award DP2 OD004613 and is supported by NIH U24 award 1U24EB023674-01 and NIH U01 award 1U01HG007708. B. Pruitt received support from American Heart Association (AHA) 1205987-120-UAKOD as well as NIH 1R21HL13099301. A. Ribeiro received support from AHA Fellowship 14POST18360018. J.C. Wu is supported by NIH R01 HL130020 and NIH R01 HL126527. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
DISCLOSURES
E. Ashley is a Founder of Personalis and DeepCell, Inc, and an advisor for SequenceBio. M. Wheeler is an equity partner in Personalis, Inc and has been a consultant to MyoKardia.
AUTHOR CONTRIBUTIONS
A.D., K.Z.-R., A.R., C.S., W.R.L., A.C.Y.C., M.W., and E.A.A. conceived and designed research; A.D., K.Z.-R., A.R., A.C.H.C., C.S., F.L., P.W.B., and W.R.L. performed experiments; A.D., K.Z.-R., A.R., A.C.H.C., and A.C.Y.C. analyzed data; A.D., A.R., C.S., A.C.Y.C., B.L.P., M.W., and E.A.A. interpreted results of experiments; A.D., A.R., and F.L. prepared figures; A.D. drafted manuscript; A.D., K.Z.-R., A.R., P.W.B., W.R.L., A.C.Y.C., B.L.P., M.W., and E.A.A. edited and revised manuscript; A.D., K.Z.-R., A.R., A.C.H.C., C.S., F.L., P.W.B., J.C.W., A.C.Y.C., B.L.P., M.W., and E.A.A. approved final version of manuscript.
REFERENCES
- 1.Adams D, Gonzalez-Duarte A, O’Riordan WD, Yang C-C, Ueda M, Kristen AV, Tournev I, Schmidt HH, Coelho T, Berk JL, Lin K-P, Vita G, Attarian S, Planté-Bordeneuve V, Mezei MM, Campistol JM, Buades J, Brannagan TH III, Kim BJ, Oh J, Parman Y, Sekijima Y, Hawkins PN, Solomon SD, Polydefkis M, Dyck PJ, Gandhi PJ, Goyal S, Chen J, Strahs AL, Nochur SV, Sweetser MT, Garg PP, Vaishnaw AK, Gollob JA, Suhr OB. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N Engl J Med 379: 11–21, 2018. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 2.Adams WJ, Pong T, Geisse NA, Sheehy SP, Diop-Frimpong B, Parker KK. Engineering design of a cardiac myocyte. J Comput Aided Mater Des 14: 19–29, 2007. doi: 10.1007/s10820-006-9045-6. [DOI] [Google Scholar]
- 3.Adhikari AS, Trivedi DV, Sarkar SS, Song D, Kooiker KB, Bernstein D, Spudich JA, Ruppel KM. Hypertrophic cardiomyopathy mutations at the folded-back sequestered β-cardiac myosin S1–S2 and S1–S1 interfaces release sequestered heads and increase myosin enzymatic activity (Preprint) bioRxiv 2019. doi: 10.1101/537159. [DOI] [PMC free article] [PubMed]
- 4.Alamo L, Ware JS, Pinto A, Gillilan RE, Seidman JG, Seidman CE, Padrón R. Effects of myosin variants on interacting—heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. eLife 6: e24634, 2017. doi: 10.7554/eLife.24634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben Jehuda R, Eisen B, Shemer Y, Mekies LN, Szantai A, Reiter I, Cui H, Guan K, Haron-Khun S, Freimark D, Sperling SR, Gherghiceanu M, Arad M, Binah O. CRISPR correction of the PRKAG2 gene mutation in the patient’s induced pluripotent stem cell-derived cardiomyocytes eliminates electrophysiological and structural abnormalities. Heart Rhythm 15: 267–276, 2018. doi: 10.1016/j.hrthm.2017.09.024. [DOI] [PubMed] [Google Scholar]
- 6.Bongianino R, Denegri M, Mazzanti A, Lodola F, Vollero A, Boncompagni S, Fasciano S, Rizzo G, Mangione D, Barbaro S, Di Fonso A, Napolitano C, Auricchio A, Protasi F, Priori SG. Allele-Specific Silencing of Mutant mRNA Rescues Ultrastructural and Arrhythmic Phenotype in Mice Carriers of the R4496C Mutation in the Ryanodine Receptor Gene (RYR2). Circ Res 121: 525–536, 2017. doi: 10.1161/CIRCRESAHA.117.310882. [DOI] [PubMed] [Google Scholar]
- 7.Bray M-A, Sheehy SP, Parker KK. Sarcomere alignment is regulated by myocyte shape. Cell Motil Cytoskeleton 65: 641–651, 2008. doi: 10.1002/cm.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke MA, Cook SA, Seidman JG, Seidman CE. Clinical and Mechanistic Insights Into the Genetics of Cardiomyopathy. J Am Coll Cardiol 68: 2871–2886, 2016. doi: 10.1016/j.jacc.2016.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll JB, Warby SC, Southwell AL, Doty CN, Greenlee S, Skotte N, Hung G, Bennett CF, Freier SM, Hayden MR. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol Ther 19: 2178–2185, 2011. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cashman TJ, Josowitz R, Johnson BV, Gelb BD, Costa KD. Human Engineered Cardiac Tissues Created Using Induced Pluripotent Stem Cells Reveal Functional Characteristics of BRAF-Mediated Hypertrophic Cardiomyopathy. PLoS One 11: e0146697, 2016. doi: 10.1371/journal.pone.0146697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drawnel FM, Boccardo S, Prummer M, Delobel F, Graff A, Weber M, Gérard R, Badi L, Kam-Thong T, Bu L, Jiang X, Hoflack J-C, Kiialainen A, Jeworutzki E, Aoyama N, Carlson C, Burcin M, Gromo G, Boehringer M, Stahlberg H, Hall BJ, Magnone MC, Kolaja K, Chien KR, Bailly J, Iacone R. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Reports 9: 810–821, 2014. doi: 10.1016/j.celrep.2014.09.055. [DOI] [PubMed] [Google Scholar]
- 12.Franck C, Maskarinec SA, Tirrell DA, Ravichandran G. Three-dimensional traction force microscopy: a new tool for quantifying cell-matrix interactions. PLoS One 6: e17833, 2011. doi: 10.1371/journal.pone.0017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garfinkel AC, Seidman JG, Seidman CE. Genetic Pathogenesis of Hypertrophic and Dilated Cardiomyopathy. Heart Fail Clin 14: 139–146, 2018. doi: 10.1016/j.hfc.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, Seidman JG. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell 62: 999–1006, 1990. doi: 10.1016/0092-8674(90)90274-I. [DOI] [PubMed] [Google Scholar]
- 15.Green EM, Wakimoto H, Anderson RL, Evanchik MJ, Gorham JM, Harrison BC, Henze M, Kawas R, Oslob JD, Rodriguez HM, Song Y, Wan W, Leinwand LA, Spudich JA, McDowell RS, Seidman JG, Seidman CE. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science 351: 617–621, 2016. doi: 10.1126/science.aad3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huebsch N, Loskill P, Mandegar MA, Marks NC, Sheehan AS, Ma Z, Mathur A, Nguyen TN, Yoo JC, Judge LM, Spencer CI, Chukka AC, Russell CR, So P-L, Conklin BR, Healy KE. Automated Video-Based Analysis of Contractility and Calcium Flux in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Cultured over Different Spatial Scales. Tissue Eng Part C Methods 21: 467–479, 2015. doi: 10.1089/ten.tec.2014.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang J, Wakimoto H, Seidman JG, Seidman CE. Allele-specific silencing of mutant Myh6 transcripts in mice suppresses hypertrophic cardiomyopathy. Science 342: 111–114, 2013. doi: 10.1126/science.1236921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karakikes I, Ameen M, Termglinchan V, Wu JC. Human induced pluripotent stem cell-derived cardiomyocytes: insights into molecular, cellular, and functional phenotypes. Circ Res 117: 80–88, 2015. doi: 10.1161/CIRCRESAHA.117.305365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim C. iPSC technology–Powerful hand for disease modeling and therapeutic screen. BMB Rep 48: 256–265, 2015. doi: 10.5483/BMBRep.2015.48.5.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, Han L, Yen M, Wang Y, Sun N, Abilez OJ, Hu S, Ebert AD, Navarrete EG, Simmons CS, Wheeler M, Pruitt B, Lewis R, Yamaguchi Y, Ashley EA, Bers DM, Robbins RC, Longaker MT, Wu JC. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell 12: 101–113, 2013. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Limpitikul WB, Dick IE, Tester DJ, Boczek NJ, Limphong P, Yang W, Choi MH, Babich J, DiSilvestre D, Kanter RJ, Tomaselli GF, Ackerman MJ, Yue DT. A Precision Medicine Approach to the Rescue of Function on Malignant Calmodulinopathic Long-QT Syndrome. Circ Res 120: 39–48, 2017. doi: 10.1161/CIRCRESAHA.116.309283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowey S, Lesko LM, Rovner AS, Hodges AR, White SL, Low RB, Rincon M, Gulick J, Robbins J. Functional effects of the hypertrophic cardiomyopathy R403Q mutation are different in an α- or β-myosin heavy chain backbone. J Biol Chem 283: 20579–20589, 2008. doi: 10.1074/jbc.M800554200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maharshi V, Hasan S. Nusinersen: The First Option Beyond Supportive Care for Spinal Muscular Atrophy. Clin Drug Investig 37: 807–817, 2017. doi: 10.1007/s40261-017-0557-5. [DOI] [PubMed] [Google Scholar]
- 24.Maron BJ, Maron MS. Hypertrophic cardiomyopathy. Lancet 381: 242–255, 2013. doi: 10.1016/S0140-6736(12)60397-3. [DOI] [PubMed] [Google Scholar]
- 25.Matsa E, Dixon JE, Medway C, Georgiou O, Patel MJ, Morgan K, Kemp PJ, Staniforth A, Mellor I, Denning C. Allele-specific RNA interference rescues the long-QT syndrome phenotype in human-induced pluripotency stem cell cardiomyocytes. Eur Heart J 35: 1078–1087, 2014. doi: 10.1093/eurheartj/eht067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montag J, Kowalski K, Makul M, Ernstberger P, Radocaj A, Beck J, Becker E, Tripathi S, Keyser B, Mühlfeld C, Wissel K, Pich A, van der Velden J, Dos Remedios CG, Perrot A, Francino A, Navarro-López F, Brenner B, Kraft T. Burst-Like Transcription of Mutant and Wildtype MYH7-Alleles as Possible Origin of Cell-to-Cell Contractile Imbalance in Hypertrophic Cardiomyopathy. Front Physiol 9: 359, 2018. doi: 10.3389/fphys.2018.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montag J, Syring M, Rose J, Weber A-L, Ernstberger P, Mayer A-K, Becker E, Keyser B, Dos Remedios C, Perrot A, van der Velden J, Francino A, Navarro-Lopez F, Ho CY, Brenner B, Kraft T. Intrinsic MYH7 expression regulation contributes to tissue level allelic imbalance in hypertrophic cardiomyopathy. J Muscle Res Cell Motil 38: 291–302, 2017. doi: 10.1007/s10974-017-9486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Musunuru K, Sheikh F, Gupta RM, Houser SR, Maher KO, Milan DJ, Terzic A, Wu JC; American Heart Association Council on Functional Genomics and Translational Biology; Council on Cardiovascular Disease in the Young; and Council on Cardiovascular and Stroke Nursing . Induced Pluripotent Stem Cells for Cardiovascular Disease Modeling and Precision Medicine: A Scientific Statement From the American Heart Association. Circ Genom Precis Med 11: e000043, 2018. doi: 10.1161/HCG.0000000000000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nag S, Sommese RF, Ujfalusi Z, Combs A, Langer S, Sutton S, Leinwand LA, Geeves MA, Ruppel KM, Spudich JA. Contractility parameters of human β-cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of motor function. Sci Adv 1: e1500511, 2015. doi: 10.1126/sciadv.1500511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelham RJ Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA 94: 13661–13665, 1997. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rape AD, Guo W-H, Wang Y-L. The regulation of traction force in relation to cell shape and focal adhesions. Biomaterials 32: 2043–2051, 2011. doi: 10.1016/j.biomaterials.2010.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ribeiro AJS, Ang Y-S, Fu J-D, Rivas RN, Mohamed TMA, Higgs GC, Srivastava D, Pruitt BL. Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proc Natl Acad Sci USA 112: 12705–12710, 2015. doi: 10.1073/pnas.1508073112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ribeiro AJS, Schwab O, Mandegar MA, Ang Y-S, Conklin BR, Srivastava D, Pruitt BL. Multi-Imaging Method to Assay the Contractile Mechanical Output of Micropatterned Human iPSC-Derived Cardiac Myocytes. Circ Res 120: 1572–1583, 2017. doi: 10.1161/CIRCRESAHA.116.310363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribeiro MC, Tertoolen LG, Guadix JA, Bellin M, Kosmidis G, D’Aniello C, Monshouwer-Kloots J, Goumans M-J, Wang Y-L, Feinberg AW, Mummery CL, Passier R. Functional maturation of human pluripotent stem cell derived cardiomyocytes in vitro–correlation between contraction force and electrophysiology. Biomaterials 51: 138–150, 2015. doi: 10.1016/j.biomaterials.2015.01.067. [DOI] [PubMed] [Google Scholar]
- 35.Rudnick SI, Swaminathan J, Sumaroka M, Liebhaber S, Gewirtz AM. Effects of local mRNA structure on posttranscriptional gene silencing. Proc Natl Acad Sci USA 105: 13787–13792, 2008. doi: 10.1073/pnas.0805781105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarkar SS, Trivedi DV, Morck MM, Adhikari AS, Pasha SN, Ruppel KM, Spudich JA. The molecular basis of hypercontractility caused by the hypertrophic cardiomyopathy mutations R403Q and R663H (Preprint) bioRxiv 2019. doi: 10.1101/543413. [DOI]
- 37.Scoles DR, Pulst SM. Oligonucleotide therapeutics in neurodegenerative diseases. RNA Biol 15: 707–714, 2018. doi: 10.1080/15476286.2018.1454812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sewanan LR, Campbell SG. Modelling sarcomeric cardiomyopathies with human cardiomyocytes derived from induced pluripotent stem cells. J Physiol JP276753, 2019. doi: 10.1113/JP276753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma A, Li G, Rajarajan K, Hamaguchi R, Burridge PW, Wu SM. Derivation of highly purified cardiomyocytes from human induced pluripotent stem cells using small molecule-modulated differentiation and subsequent glucose starvation. J Vis Exp (97): 52628, 2015. doi: 10.3791/52628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skotte NH, Southwell AL, Østergaard ME, Carroll JB, Warby SC, Doty CN, Petoukhov E, Vaid K, Kordasiewicz H, Watt AT, Freier SM, Hung G, Seth PP, Bennett CF, Swayze EE, Hayden MR. Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: providing a therapeutic option for all Huntington disease patients. PLoS One 9: e107434, 2014. doi: 10.1371/journal.pone.0107434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith AST, Macadangdang J, Leung W, Laflamme MA, Kim D-H. Human iPSC-derived cardiomyocytes and tissue engineering strategies for disease modeling and drug screening. Biotechnol Adv 35: 77–94, 2017. doi: 10.1016/j.biotechadv.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Southwell AL, Skotte NH, Kordasiewicz HB, Østergaard ME, Watt AT, Carroll JB, Doty CN, Villanueva EB, Petoukhov E, Vaid K, Xie Y, Freier SM, Swayze EE, Seth PP, Bennett CF, Hayden MR. In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol Ther 22: 2093–2106, 2014. doi: 10.1038/mt.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tripathi S, Schultz I, Becker E, Montag J, Borchert B, Francino A, Navarro-Lopez F, Perrot A, Özcelik C, Osterziel K-J, McKenna WJ, Brenner B, Kraft T. Unequal allelic expression of wild-type and mutated β-myosin in familial hypertrophic cardiomyopathy. Basic Res Cardiol 106: 1041–1055, 2011. doi: 10.1007/s00395-011-0205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Unverferth DV, Baker PB, Pearce LI, Lautman J, Roberts WC. Regional myocyte hypertrophy and increased interstitial myocardial fibrosis in hypertrophic cardiomyopathy. Am J Cardiol 59: 932–936, 1987. doi: 10.1016/0002-9149(87)91128-3. [DOI] [PubMed] [Google Scholar]
- 45.Wheeler M, Ashley EA, Zaleta-Rivera KM. Oligonucleotides and Methods for Treatment of Cardiomyopathy Using RNA Interference [Online]. US Patent: 2016. https://patentimages.storage.googleapis.com/d1/60/31/a7ac28e276753a/US20160348103A1.pdf [16 Apr. 2019].
- 46.Wheelwright M, Win Z, Mikkila JL, Amen KY, Alford PW, Metzger JM. Investigation of human iPSC-derived cardiac myocyte functional maturation by single cell traction force microscopy. PLoS One 13: e0194909, 2018. doi: 10.1371/journal.pone.0194909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida T, Naito Y, Sasaki K, Uchida E, Sato Y, Naito M, Kawanishi T, Obika S, Inoue T. Estimated number of off-target candidate sites for antisense oligonucleotides in human mRNA sequences. Genes Cells 23: 448–455, 2018. doi: 10.1111/gtc.12587. [DOI] [PubMed] [Google Scholar]
- 48.Zaleta K, Dainis A, Ribeiro AJS, Sanchez-Cordero P, Rubio G, Shang C, Liu J, Finsterbach T, Sinha N, Jain N, Hajjar R, Kay MA, Szczesna-Corday D, Pruitt BL, Wheeler MT, Ashley EA. Allele-specific silencing ameliorates restrictive cardiomyopathy due to a human myosin regulatory light chain mutation (Preprint) bioRxiv 2019. doi: 10.1101/559468. [DOI] [PubMed]