Fig. 3.
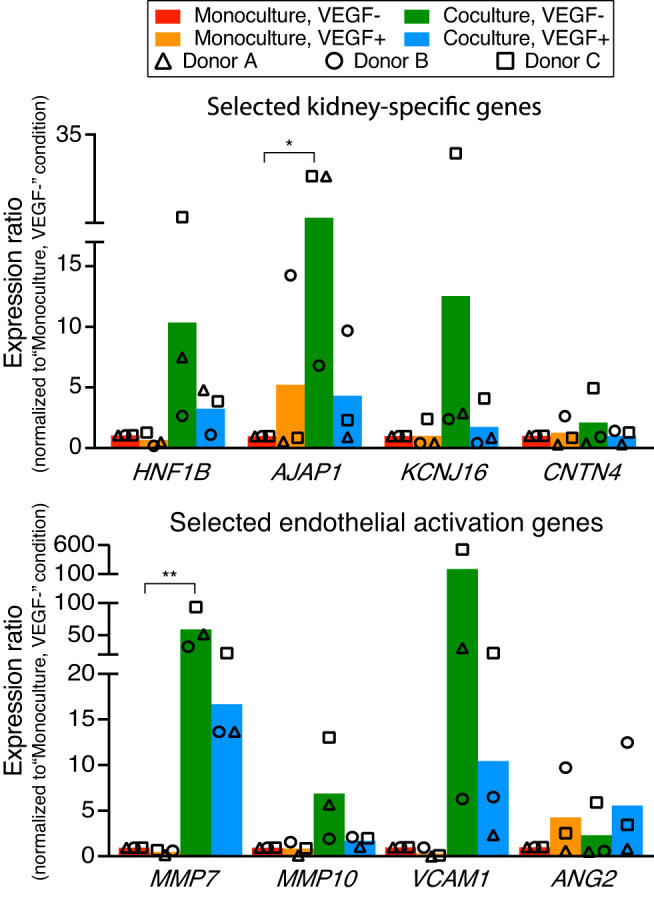
Transcriptional changes of human kidney peritubular microvascular ECs (HKMECs) by coculturing with human kidney proximal tubule epithelial cells (HPTECs). Quantitative RT-PCR of selected HKMEC-specific genes (top) and activation genes (bottom) showed upregulation in the “coculture, VEGF−” condition (green) compared with the “monoculture, VEGF−” condition (red). Each plotted point represents data from an independent human donor (pooled from 5 replicate microculture devices per donor, average of three quantitative RT-PCR technical replicates), with each normalized to the “monoculture, VEGF−” condition (which was set to 1). Each colored bar represents the average ratio of the three donors. Statistical comparisons (as described in materials and methods) are shown for genes showing significant differences between “coculture, VEGF−” and “monoculture, VEGF−” as described in materials and methods. *P ≤ 0.05; **P ≤ 0.01. HNFB1, hepatocyte nuclear factor 1 homeobox B; AJAP1, adherens junctions-associated protein 1; KCNJ16, potassium voltage-gated channel subfamily J member 16; CNTN4, contactin 4; MMP7, matrix metalloproteinase-7; MMP10, matrix metalloproteinase-10; VCAM1, vascular cell adhesion molecule-1; ANG2, angiopoietin 2.
