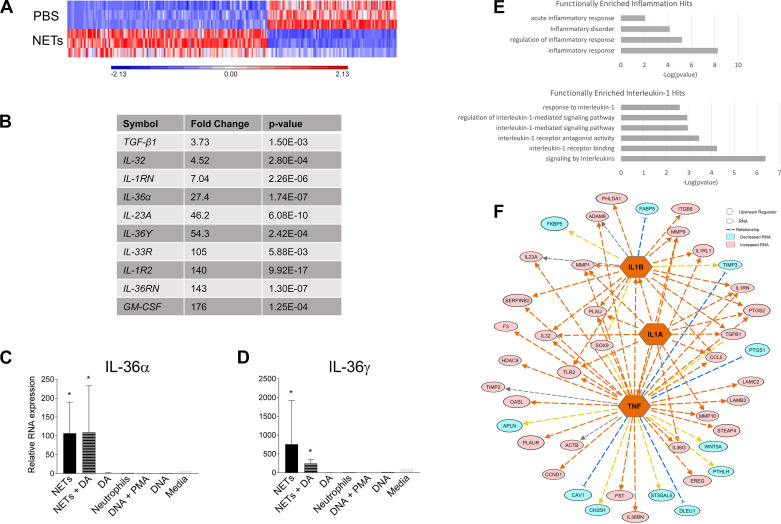
Fig. 4.
Neutrophil extracellular traps (NETs) influence gene expression in human bronchial epithelial cells (HBEs). HBEs grown at air-liquid interface were exposed to PBS, 5 μg/mL NETs, 5 μg/mL NETs + 0.5 μg/mL dornase alfa (DA), or 0.5 μg/mL DA for 18 h in triplicate wells. Differential gene expression was assessed by RNA sequencing. A: expression of 150 RNAs was increased, whereas expression of 97 RNAs was decreased, after exposure to 5 μg/mL NETs compared with PBS (P < 0.01 and a fold change >2.5). B: the most highly induced RNAs in response to NETs, particularly IL-1 family members (IL-36α, IL-36γ, etc.) and GM-CSF. C and D: HBEs were exposed to the above conditions as well as unstimulated human neutrophils, 0.5 µg/mL genomic DNA (gDNA) + 500 nM PMA, 0.5 µg/mL gDNA, or media control for 18 h. IL-36α and IL-36γ gene expression was assessed by RT-PCR, normalized to 18S, and analyzed by one-way ANOVA comparing each condition relative to RNA expression in HBEs exposed to PBS. Measured in biologic triplicate wells and run in experimental triplicates. E: functional enrichment analysis using ToppGene indicated that IL-1 cytokines and their corresponding antagonists were increased from all three HBE donors. F: schematic representation of the upstream regulators (orange) predicted by Ingenuity Pathway Analysis to influence the transcriptional changes seen in HBEs exposed to NETs. Pink indicates increased expression of RNAs, and aqua indicates decreased expression of RNAs. (HBE donors = 3, NET donors = 3.) *P < 0.05.