Abstract
We detail a heterobifunctional, 7-aminocoumarin photocleavable (PC) linker with unique properties to covalently attach Abs to surfaces and subsequently release them with visible light (400–450 nm). The PC linker allowed rapid (2 min) and efficient (>90%) release of CTCs and EVs without damaging their molecular cargo.
Liquid biopsies consist of disease-associated markers harvested from body fluids that can be secured in a minimally invasive manner to provide information for guiding patient treatment (i.e., precision medicine) by securing molecular characteristics of the disease.1 Initially focused on epithelial cancers, liquid biopsies have been extended to other diseases such as blood cancers and stroke.2, 3 Common liquid biopsy markers include, but are not limited to, nanometer-sized extracellular vesicles, EVs,4 and cells (circulating tumour cells, CTCs,2 or CD8(+) T-cells for stroke5).
Microfluidics that use affinity-agents (e.g., Abs) attached to their surfaces can specifically enrich disease-associated EVs1 or CTCs2 from biological samples. Further, affinity-enrichment can fractionate different marker subsets, such as targeting epithelial CTCs via EpCAM (epithelial cell adhesion molecule) and mesenchymal CTCs via FAPα (fibroblast activation protein alpha).6
While early studies focused on biomarker enumeration to indicate disease status, the Precision Medicine initiative now requires profiling the disease’s molecular composition.7 Thus, there is a need to integrate enrichment with advanced molecular profiling.2, 5, 8 For nanometer-sized EVs, enumeration requires off-line nanoparticle tracking analysis (NTA) and transmission electron microscopy (TEM) for enumeration,9 data critical for mRNA expression analyses.10 Thus, it is necessary to release affinity-enriched markers without damaging the marker or its cargo.
New chemistries have been developed for “catch and release,” where biomarkers are affinity-enriched on a solid-phase (i.e., microfluidic device) then released for analyses.11, 12 Release has been accomplished using degraded polymer coatings, proteolytically digested Abs, or cleaved Ab linkers with enzymes or UV light.2 Performance metrics for CTC catch and release are: (i) Recovery (CTC yield/input); (ii) purity (CTCs/total cell count); (iii) release efficiency (CTCs released/recovered); (iv) cell viability; (v) preserving molecular cargo; and (vi) minimizing assay cost, workflow, and instrumentation.
Previously, we reported a single-stranded oligonucleotide linker for Ab immobilization and subsequent enzymatic release. Enzymatic cleavage of a dU nucleotide in the linker released >90% of CTCs, preserved viability (>85%), and enabled off-line immunophenotyping and cytogenetic analyses.13 We incorporated the oligonucleotide linker into a sinusoidal microfluidic chip that achieved >75% CTC recovery from clinical samples, high purity (>90%)13 and 80–100% test positivity for several epithelial cancers.2, 6 We also used the oligonucleotide linker to enrich leukemic cells3 and immune cells responding to inflammatory processes associated with acute ischemic stroke (AIS).5 However, the reaction time (~60 min) was prohibitively long for time-sensitive analyses as required for AIS diagnosis, where the therapeutic time window for recombinant tissue plasminogen activator (r-tPA) treatment is ~4.5 h.5
We report a photocleavable 7-amino coumarin Ab linker for specific biomarker enrichment and reagent-less release (catch and release). 7- amino coumarin was used previously as a photolabile group to release different targets (drugs,14, 15 thiol bearing proteins16 and CTCs17) over a wide wavelength range (UV, visible, and near-IR), but UV induced DNA/RNA damage, low release efficiencies (50–80%) and lengthy release times (~120 min) limited their use. Here, we synthesized a 7-(diethylamino)coumaryl-4-methyl photorelease agent18 (see Scheme S1 and SI for synthesis of the PC linker and structural characterization) that rapidly cleaves (Figure S1A) with visible light (400–450 nm,19 quantum efficiency of coumarin dye is 0.2518) minimizing nucleic acid damage and little to no side reactions (Scheme 1). Also, minimal changes in the UV/vis (Figure S1B) and fluorescence emission (Figure S1C) as a function of cleavage were observed. The PC linker is unique in its structure; the PC linker contains amino and carboxy termini (i.e., amino acid) to allow for two well-established EDC/NHS reactions to first covalently attach the linker to a carboxylated surface, and then to an Ab.
Scheme 1.
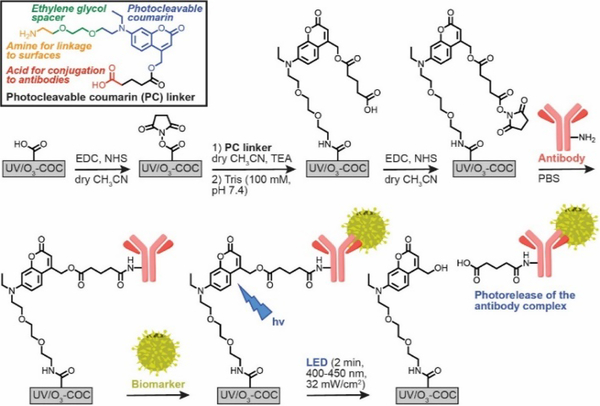
Structure of the heterobifunctional PC linker and immobilization strategy employing two EDC/NHS coupling reactions. The PC linker is immobilized to carboxylated (UV/O3-oxidized) surfaces via the linker’s primary amine group. Abs are then anchored to the linker’s -COOH handle. After biomarker purification, a visible LED (400–450 nm) cleaves the PC linker. Immobilization reactions are conducted in organic solvent to mitigate NHS ester hydrolysis, which could lead to direct (and non-releasable) Ab conjugation to the -COOH surface.
We will demonstrate efficient and rapid release with inexpensive LEDs with minimal effects on the marker and its molecular cargo. Also, we show that the PC linker can be used for the catch and release of CTCs and EVs with no to minimal damage to their molecular cargo.
The immobilization chemistry was designed to allow for the facile surface attachment of a recognition element, for example an Ab, used in a microfluidic device, which in this case was fabricated in cyclic olefin copolymer (COC) for enriching rare liquid biopsy markers. COC devices can be mass produced by injection moulding and photo-activated using UV/O3 irradiation to yield surface-confined carboxylic acid (-COOH) scaffolds.20 The PC linker contains a primary amine with an ethylene glycol spacer for EDC/NHS coupling to the surface -COOH groups, and the linker’s opposing -COOH covalently anchors Abs via a second EDC/NHS reaction. Solid-phase conjugation prevents PC linker cross-linking, and identical reaction chemistry simplifies synthetic preparation and subsequent derivatization reactions. However, EDC/NHS reactions in aqueous buffers can result in NHS ester hydrolysis. The resulting surface -COOH groups can directly attach Abs to the surface during the second EDC/NHS reaction. These directly-attached Abs would not allow photorelease of the enriched liquid biopsy marker. To mitigate NHS ester hydrolysis, we investigated the ability to perform EDC/NHS reactions in an anhydrous solvent, such as acetonitrile (ACN).
COC is stable in many organic solvents, but its stability is unknown following UV/O3 activation, which generates various oxidation products and polymer fragments.20 Surface analyses (Table S1) of UV/O3-activated COC exposed to ACN showed increased hydrophobicity (64.7° water contact angle vs 56.7° in buffer), ~3-fold lower -COOH surface densities, but constant ATR-FTIR oxidation signals. Loss of –COOH groups at the surface was presumably due to solubilization of oxidized polymer fragments.
We tested the immobilization efficiency using a 3’ Cy5-labeled oligonucleotide reporter bearing a primary amine on its 5’ end to UV/O3-COC surfaces pre-treated with ACN or with MES buffer (pH 4.8). ACN treatment yielded oligonucleotide reporter surface coverages comparable to MES buffer treatment (Figure S2A). However, improved reaction efficiency was observed when conducting the EDC/NHS coupling reaction in ACN (Figure S2B) likely due to eliminating NHS ester hydrolysis.
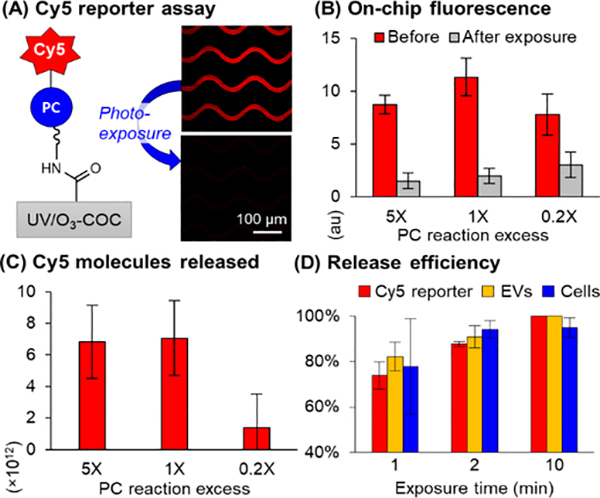
We next immobilized the PC linker (in ACN and TEA) to UV/O3-activated COC microfluidic devices and measured the immobilization efficiency using the Cy5-labeled oligonucleotide. TEA was used with dry ACN to facilitate amide bond formation between the primary amine and NHS ester. Unreacted NHS ester was quenched with Tris base to prevent direct attachment of Cy5-labeled oligonucleotides. Then, we reacted the linker’s -COOH terminus with EDC/NHS in ACN, and immobilized Cy5-labeled oligonucelotides in buffer using the PC linker (Figure 1A). We then used an LED (λmax = 412 nm, 32 ±4 mW/cm2, Figure S3) to cleave the PC linker. Decreasing on-chip fluorescence of the Cy5-labeled oligonucleotide (Figure 1A and 1B) was confirmatory for successful release through controls (Figure S4) and quantification of Cy5-labeled oligonucleotides in the off-chip effluent by fluorescence spectroscopy (Figure 1C). The remaining on-chip fluorescence may be due to autofluorescence from the UV/O3 activated surface and/or non-released Cy5-labeled oligonucleotides.
Figure 1.
(A) Cy5-oligonucleotide reporters were immobilized via the PC linker at 0.2–5X of the theoretical monolayer reaction excess (0.11–2.65 mM in ACN) considering the microfluidic device’s surface area. (B) On-chip microscopy (n= 3) and (C) fluorescence spectroscopy of released Cy5 molecules (n = 3) show saturation at 1–5X. (D) Cy5-labeled oligonucleotides (n = 3), EVs (n=5), and SKBR3 cells (n=3) were released with 88%, 91%, and 94% efficiency in 2 min, respectively.
The PC linker concentration required to bind the maximum number of Abs was required to maximize recovery of the target. For that, we varied the PC linker reaction excess compared to a monolayer (considering surface area) and observed the same number of Cy5-labeled reporters released in 1–5X PC linker excess (Figure 1C). Thus, we could saturate the device at 1X PC linker reaction excess (0.56 nmol/cm2, 1.82 × 1015 molecules per device) and the PC linker concentration was sufficient to maximize the recovery. We varied LED exposure time and released 74 ±6% Cy5-labeled oligonucleotides in 1 min and 88 ±1% release efficiency in 2 min, Figure 1D, resulting in release of 6.4 ×1012 molecules (Figure 1C).
The PC linker was used to immobilize anti-EpCAM Abs in a microfluidic device for CTC affinity-enrichment.6 EpCAM(+) SKBR3 cells were spiked into healthy donor blood (69–269 SKBR3 cells/mL) to evaluate the assay performance with PC linker. We pre-stained SKBR3 cells with Hoechst dye and, after enrichment, stained all cells with SYTO 82, a membrane-permeable nuclear dye that is spectrally distinct from Hoechst. SKBR3 cells were dual-stained, while leukocytes were only stained with SYTO 82 enabling the determination of recovery by a self-referencing method13 and purity of the enriched fraction (Figure 2A).
Figure 2.
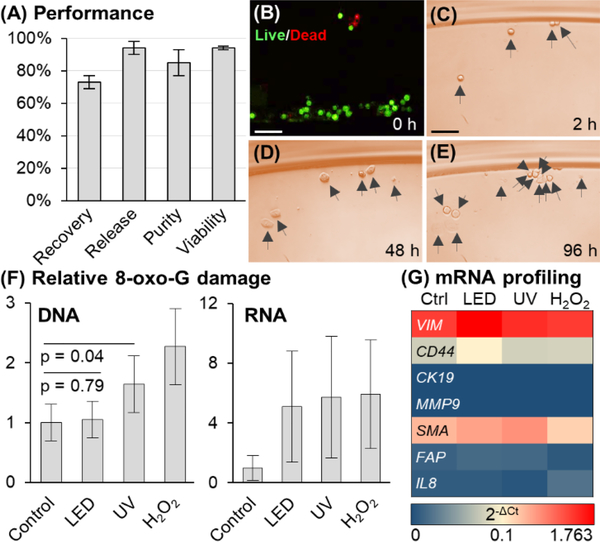
(A) Performance of sinusoidal microfluidic device using PC linker for anti-EpCAM enrichment of SKBR3 cells spiked into whole blood (N = 3). (B) LED release had no effect on viability, and (C–E) released cells in culture for 2–96 h (Scale bars = 100 μm). (F) DNA/RNA oxidative damage (N = 3) assessed for 2 min LED exposure, equivalent UV dose (18.5 J), and 300 μM H2O2 (30 min) of Hs578T cells. DNA and RNA damage is normalized to control. (32.2 pg 8-oxo-G per 400 ng DNA and 7.15 pg 8-oxo-G per 400 ng RNA). The DNA-derived ELISA 8-oxo-G calibration curve could not quantify RNA damage absolutely. (G) mRNA profiling by RT-qPCR (N = 3) of Hs578T cells following no irradiation, LED or UV light exposure, or H2O2 treatment.
We next used the PC linker to immobilize anti-EpCAM Abs in a sinusoidal microfluidic device for CTC affinity-enrichment.6 We targeted SKBR3 cells spiked into blood (50–27069-269 SKBR3 cells/mL) to evaluate possible interferences of the sample matrix on the PC linker’ performance. We pre-stained SKBR3 cells with Hoechst dye and, after enrichment, stained all cells with SYTO 82, a membrane-permeable nuclear dye that is spectrally distinct from Hoechst. SKBR3 cells were dual-stained, while leukocytes were only stained with SYTO 82 enabling the determination of the PC linker’s performance metrics of recovery using a self-referencing method13 and purity of the enriched fraction (Figure 2A).
SKBR3 cells were enriched with 85 ±8% purity (16–38 leukocytes/mL) and 73 ±4% recovery (47–202 cells), slightly lower than found for the dU linker (85 ±4%) and direct Ab attachment (96 ±12%).13 We also observed ~40% fewer Cy5-labeled oligonucleotides immobilized through the PC linker compared to direct conjugation to the surface.13 We suspect that Ab load and CTC recovery may increase if the PC linker contained a longer PEG spacer, which would allow for more access to targets and potentially less surface crowding. We used the PC linker to immobilize IgG 2A isotype Ab to evaluate nonspecific cell recover, which was 3 ±2%. The release of SKBR3 cells was rapid with 94 ±4% efficiency after 2 min of LED exposure (Figure 1D). Other breast cancer cell lines were also enriched and released with 88 ±10% and 91 ±4% release efficiency for MCF7 and Hs578T cells, respectively. We also found that the release efficiency was independent of cell antigen expression (Figure S5A for flow cytometry results and correlation to release efficiency, Figure S5B).
After exposure to visible LED light, 94 ±1% of released SKBR3 cells were viable, the same as controls (Figure 2B), and could be propagated in culture for 96 h (Figure 2C – E). Released MCF7 and Hs578T cells had 96 ±6% and 99 ±3% cell viability, respectively.
However, UV irradiation can damage nucleic acids through photo-absorption and oxidation (8-oxoguanine, 8-oxo-G, production). To assess that, we measured 8-oxo-G levels in RNA and DNA for Hs578T cells exposed to visible LED and UV light (both at a dose of 18.5 J) and compared to H2O2, which is known to generate oxidative damage in DNA and RNA via oxygen free radicals. DNA damage was not detected for LED exposure but was present with UV irradiation (Figure 2F). Both exposures generated 8-oxo-G damage in RNA at comparable levels to that of H2O2 (Figure 2F). Single-stranded RNA is easily oxidized so as to protect genomic DNA from damage21 and subsequent mutations through imperfect repair pathways.22 A gene panel consisting of mesenchymal and EMT markers were selected to determine the impact of mRNA oxidative damage (Figure 2G; see Table S2 for primers used for the RT-qPCR). The mRNA expression was similar in all 3 treatments suggesting that while the 8-oxo-G damage is observed in RNA, the frequency was so low that it did not appear to alter the mRNA expression for the selected gene panel. Collectively, visible LED exposure did not affect mRNA expression or cause oxidative DNA damage, whereas UV irradiation induced DNA 8-oxo-G damage. Such DNA damage could cause false positives for clinical analysis, especially at the single cell level, which is commonly encountered when analyzing CTCs.
We also evaluated the impact of attached Abs of enriched cells in terms of mRNA expression following photorelease using a series of stress genes. Affinity isolated SKBR3 cells were released, and mRNA expression was analysed via RT-qPCR. We observed statistically similar mRNA expression profiles (Figure S6) in both released and cells not containing attached Abs.
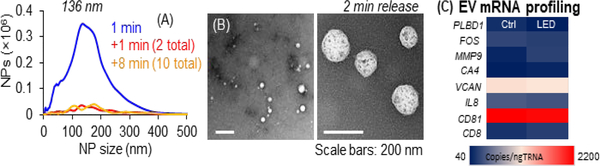
Lastly, we tested the PC linker for EV catch and release. Our group recently affinity enriched CD8(+) EVs as a liquid biopsy marker to diagnose AIS. We relied on the dU linker strategy to release EVs,23 but the 60 min enzymatic reaction increased total assay time, approaching the ~4.5 h therapeutic time window. Thus, we investigated the use of the PC linker to reduce processing time for releasing AIS-associated EVs. We immobilized the PC linker and anti-CD8 Abs to a UV/O3-COC microfluidic device specifically-designed to enrich EVs with high efficiency.23 The expression of CD8 antigen in MOLT-3 cells was reported as 13.5%, thus the MOLT-3 cell line was used as a model for these studies.24 CD8(+) EVs were affinity selected from MOLT-3 conditioned media and photoreleased for NTA and TEM analysis. From the MOLT-3 conditioned media, we enriched 8.2 ±0.2 × 107 nanoparticles (NPs) with an EV size of ~136 nm, similar to TEM (Figure 3A, B). EV release was rapid and efficient; 82 ±6% of EVs released after 1 min LED exposure, and 91 ±5% released within 2 min of exposure (Figure 1D). The PC linker’s rapid cleavage reduces AIS assay workflow by >58 min compared to enzymatic release.
Figure 3.
EVs affinity-enrichment (anti-CD8 mAbs). The EVs were enriched from MOLT-3 conditioned media and released by LED exposure from the EV enrichment microfluidic device. The released EVs were subjected to NTA (A), TEM (B), and ddPCR (C). The ddPCR was carried out on 8 genes. Among them, five genes (PLBD1, FOS, MMP9, CA4 and VCAN) are known to be dysregulated as a result of AIS event. See Table S3 for the sequences of the primers used for the ddPCR.
Further, the EVs mRNA gene profile was obtained by droplet digital PCR (see Table S3 for primer sequences) after 2 min of LED exposure and compared with control EVs (not exposed to blue light). The gene panel consisted of genes dysregulated as a result of AIS and stress genes (i.e., PLBD1, FOS, MMP9, CA4, VCAN and IL8). CD81 and CD8 genes are EV-specific and used to confirm the selected EV markers. A strong positive correlation (0.99) of the two data sets (Figure S7) implied 2 min of LED exposure did not significantly affect EV-mRNA cargo integrity (Figure 3C).
We successfully demonstrated a PC linker for the “catch and release” of clinically-relevant liquid biopsy markers (CTCs and EVs) attached to a –COOH surface using two-step EDC/NHS coupling chemistry. The PC linker is easily adaptable for any affinity agent bearing a primary amine, such as Abs and aptamers, as well as different microfluidic platforms. Elements in biologically complex matrices, such as blood, did not interfere or affect the assay as judged by high cell recovery (>70%). We also showed high cell viability and the possibility of culturing released CTCs. Unlike UV exposure, our PC linker cleaves in response to visible light, and thus, does not affect DNA integrity. Importantly, our PC linker was able to release enriched liquid biopsy markers efficiently (>90%) and rapidly (2 min) with a blue LED (400–450 nm). AIS diagnostics takes full advantage of the PC linker’s rapid release to reduced assay time for diagnostic tests that possess short therapeutic time windows. This reagent-free release method is inexpensive and well-suited for clinical settings because it obviates the need for thermally mediated enzymatic reactions. While ambient light can cause photocleavage, once the enrichment device has been loaded with the affinity agent using the PC linker, the device can be wrapped in a rubylith film to protect the integrity of the PC linker (see Figure S2A).
Supplementary Material
Acknowledgments
The authors would like to thank the NIH for funding of this work (NIBIB: P41 EB020594; NCI: P30 CA168524, R01 CA211720; NIGMS: P20 GM130423, P20GM103638). We also acknowledge the KU Microscopy and Analytical Imaging Laboratory for TEM imaging (DOD-47040-2974000-908), KU Endowment Funds (RSG). We thank the Biospecimen Repository Core for providing healthy donor blood.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Electronic Supporting Information (ESI)
ESI includes experimental methods: PC linker synthesis and characterization; and immobilization; LED light exposure system; PC release effect on DNA damage; mRNA expression analysis; and results: synthesis of PC linker, Scheme S1; photocleavage of PC linker, Figure S1; anhydrous solvent effects, Table S1; LED exposure system, Figure S2; ACN vs. MES buffer EDC/NHS reactions, Figure S3; Cy5 reporter assay, Figure S4; flow cytometry of cell lines, Figure S5; Figure S6 gene expression of Hs578T and SKBR3 cells, Table S2; MOLT-3 EV mRNA profile, Table S3).
References
- 1.Campos CDM, Jackson JM, Witek MA and Soper SA, Cancer J, 2018, 24, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson JM, Witek MA, Kamande JW and Soper SA, Chem Soc Rev, 2017, 46, 4245–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson JM, Taylor JB, Witek MA, Hunsucker SA, Waugh JP, Fedoriw Y, Shea TC, Soper SA and Armistead PM, Analyst, 2016, 141, 640–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Contreras-Naranjo JC, Wu HJ and Ugaz VM, Lab Chip, 2017, 17, 3558–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pullagurla SR, Witek MA, Jackson JM, Lindell MA, Hupert ML, Nesterova IV, Baird AE and Soper SA, Anal Chem, 2014, 86, 4058–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witek MA, Aufforth RD, Wang H, Kamande JW, Jackson JM, Pullagurla SR, Hupert ML, Usary J, Wysham WZ, Hilliard D, Montgomery S, Bae-Jump V, Carey LA, Gehrig PA, Milowsky MI, Perou CM, Soper JT, Whang YE, Yeh JJ, Martin G and Soper SA, Nat. Prec. Onc, 2017, 1, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashley EA, Nat. Rev. Genet, 2016, 17, 507–522. [DOI] [PubMed] [Google Scholar]
- 8.Sheng W, Ogunwobi OO, Chen T, Zhang J, George TJ, Liu C and Fan ZH, Lab on a Chip, 2014, 14, 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW and Thery C, J. Extracell. Vesicles, 2014, 3, 26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS and Tewari M, Proc. Natl. Acad. Sci. U. S. A, 2014, 111, 14888–14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ao Z, Parasido E, Rawal S, Williams A, Schlegel R, Liu S, Albanese C, Cote RJ, Agarwal A and Datar RH, Lab Chip, 2015, 15, 4277–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reategui E, Aceto N, Lim EJ, Sullivan JP, Jensen AE, Zeinali M, Martel JM, Aranyosi AJ, Li W, Castleberry S, Bardia A, Sequist LV, Haber DA, Maheswaran S, Hammond PT, Toner M and Stott SL, Advanced materials, 2015, 27, 1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nair SV, Witek MA, Jackson JM, Lindell MA, Hunsucker SA, Sapp T, Perry CE, Hupert ML, Bae-Jump V, Gehrig PA, Wysham WZ, Armistead PM, Voorhees P and Soper SA, Chem. Commun, 2015, 51, 3266–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong J, Xun Z, Zeng Y, Yu T, Han Y, Chen J, Li YY, Yang G and Li Y, Chemistry–A European Journal, 2013, 19, 7931–7936. [DOI] [PubMed] [Google Scholar]
- 15.Huang Q, Bao C, Ji W, Wang Q and Zhu L, J. Mat. Chem, 2012, 22, 18275–18282. [Google Scholar]
- 16.Lin Q, Bao C, Cheng S, Yang Y, Ji W and Zhu L, J Am Chem Soc, 2012, 134, 5052–5055. [DOI] [PubMed] [Google Scholar]
- 17.Lv S-W, Wang J, Xie M, Lu N-N, Li Z, Yan X-W, Cai S-L, Zhang P-A, Dong W-G and Huang W-H, Chem. Sci, 2015, 6, 6432–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin Q, Bao C, Cheng S, Yang Y, Ji W and Zhu L, J. Am. Chem. Soc, 2012, 134, 5052–5055. [DOI] [PubMed] [Google Scholar]
- 19.Givens RS, Rubina M and Wirz J, Photochem Photobiol Sci, 2012, 11, 472–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson JM, Witek MA, Hupert ML, Brady C, Pullagurla S, Kamande J, Aufforth RD, Tignanelli CJ, Torphy RJ, Yeh JJ and Soper SA, Lab Chip, 2014, 14, 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radak Z and Boldogh I, Free Radic. Biol. Med, 2010, 49, 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yasui M, Kanemaru Y, Kamoshita N, Suzuki T, Arakawa T and Honma M, DNA Repair, 2014, 15, 11–20. [DOI] [PubMed] [Google Scholar]
- 23.Wijerathne H, Witek MA, Jackson JM, Hupert ML, Baird AE and Soper SA, Submitted for publication, 2019. [Google Scholar]
- 24.Greenberg JM, Gonzalez-Sarmiento R, Arthur DC, Wilkowski CW, Streifel B and Kersey J, Blood, 1988, 72, 1755–1760. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.