Abstract
The cytosolic sulfotransferases (SULTs) are phase II conjugating enzymes that catalyze the transfer of a sulfonate group from the universal sulfate donor 3′-phosphoadenosine-5′-phosphosulfate to a nucleophilic group of their substrates to generate hydrophilic products. Sulfation has a major effect on the chemical and functional homeostasis of substrate chemicals. SULTs are widely expressed in metabolically active or hormonally responsive tissues, including the liver and many extrahepatic tissues. The expression of SULTs exhibits isoform-, tissue-, sex-, and development-specific regulations. SULTs display a broad range of substrates including xenobiotics and endobiotics. The expression of SULTs has been shown to be transcriptionally regulated by members of the nuclear receptor superfamily, such as the peroxisome proliferator–activated receptors, pregnane X receptor, constitutive androstane receptor, vitamin D receptor, liver X receptors, farnesoid X receptor, retinoid-related orphan receptors, estrogen-related receptors, and hepatocyte nuclear factor 4α. These nuclear receptors can be activated by numerous xenobiotics and endobiotics, such as fatty acids, bile acids, and oxysterols, many of which are substrates of SULTs. Due to their metabolism of xenobiotics and endobiotics, SULTs and their regulations are implicated in the pathogenesis of many diseases. This review is aimed to summarize the central role of major SULTs, including the SULT1 and SULT2 subfamilies, in the pathophysiology of liver and liver-related diseases.
SIGNIFICANCE STATEMENT
Sulfotransferases (SULTs) are indispensable in the homeostasis of xenobiotics and endobiotics. Knowing SULTs and their regulations are implicated in human diseases, it is hoped that genetic or pharmacological manipulations of the expression and/or activity of SULTs can be used to affect the clinical outcome of diseases.
Introduction
Sulfate conjugation (sulfation or sulfonation) is a major conjugating pathway responsible for the deactivation, detoxification, and excretion of xenobiotics and endogenous molecules (Falany, 1991). Sulfoconjugation was first recognized as an important metabolic pathway by Baumann (1876). At the chemical level, the cytosolic sulfotransferases (SULTs) catalyze the transfer of a negatively charged sulfonate group (SO3−) from the universal sulfate donor 3′-phosphoadenosine-5′-phosphosulfate (PAPS) onto a nucleophilic group of their substrates to generate hydrophilic products, which often promote the urinary excretion of the substrates. The high-energy sulfate donor PAPS can be generated by PAPS synthases, including PAPSS1 and PAPSS2, as well as by ATP sulfurylase and two forms of adenosine 5ʹ-phosphosulfate kinase (Mueller et al., 2018).
SULTs are widely expressed in the liver, as well as metabolically active or hormonally responsive extrahepatic tissues (Dooley et al., 2000; Gamage et al., 2006). This large family of enzymes is responsible for sulfating a variety of endogenous and exogenous molecules, including pharmaceuticals, procarcinogens, hormones, and neurotransmitters, as well as intermediates of endogenous metabolism (Dooley et al., 2000; Glatt et al., 2001; Negishi et al., 2001; Jancova et al., 2010). Sulfation often results in the inactivation of the substrates or reduced potency of ligands (Strott, 2002; Bjerregaard-Olesen et al., 2015) but with some exceptions. Their abundance in the liver and wide range of substrates suggest SULTs may act as important mediators for the development of liver diseases, such as hepatocellular carcinoma (Xie et al., 2017; Zou et al., 2017), liver fibrosis (Hardwick et al., 2013; Krattinger et al., 2016; Yetti et al., 2018), and drug-induced liver injury (Fang et al., 2016). Therefore, in this review we focus on the roles of the human and rodent SULTs in liver diseases.
The Superfamily of Sulfotransferases
The SULTs superfamily contains 62 human SULT genes and 46 murine homologs as of 2016 (Mueller et al., 2015; Herrero et al., 2016). Among SULTs, the aryl-sulfotransferase (SULT1) and the hydroxysteroid sulfotransferase (SULT2) families are two principal subfamilies of SULTs that are the major contributors to the sulfonation of many xenobiotics, including pharmaceuticals and procarcinogens, and endobiotics, including steroids, thyroid, and neurotransmitters (Kauffman, 2004; Reinen and Vermeulen, 2015).
The SULT1 family comprises five isoforms: phenol-sulfotransferases (SULT1A1/2) (Raftogianis et al., 1999); catecholamine phenol sulfotransferase (SULT1A3/4), which is only present in humans (Zou et al., 2017); thyroid hormone sulfotransferase (SULT1B1) (Saeki et al., 1998); iodothyronine sulfotransferase (SULT1C2/4) (Dubaisi et al., 2019); and estrogen sulfotransferase (EST/SULT1E1) (Zhang et al., 1998; Guo et al., 2015). The SULT2 family has two isoforms: alcohol/hydroxysteroid sulfotransferase (SULT2A1), which sulfonates hydroxysteroids (Mueller et al., 2018), such as androgens, estrogens at both the 3- and 17- positions (Ambadapadi et al., 2017), and bile acids (Huang et al., 2010), and SULT2B1b, which has a greater selectivity for 3-hydroxysteroids, such as cholesterol (Bi et al., 2018), but not for bile acids.
In addition to the SULT1 and SULT2 families of enzymes, the human genome contains two more sulfotransferase gene families, SULT4 and SULT6, encoding enzymes including SULT4A1, which is a brain-specific sulfotransferase associated with antipsychotic treatment response (Wang et al., 2014), and SULT6B1, whose physiologic function is largely unknown.
SULT1
SULT1A1/2
SULT1A1/2 in Liver Cancers.
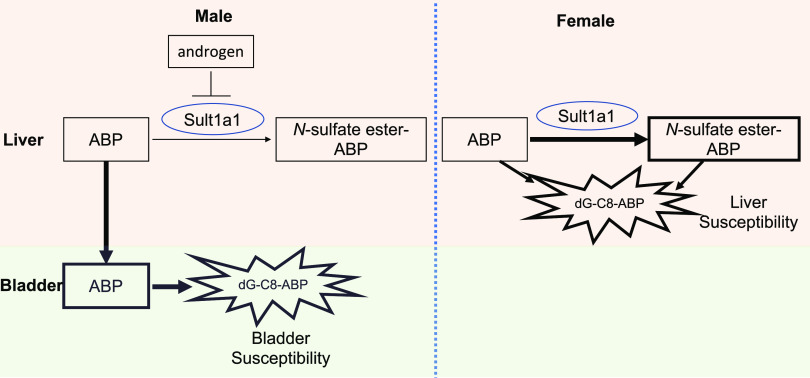
The human SULT1A1 and SULT1A2, also known as aryl/phenol or thermostable sulfotransferases, exhibit a broad substrate range with specificity for phenolic compounds (Raftogianis et al., 1997, 1999). SULT1A1 is widely distributed throughout the body with high abundance in the liver, lung, brain, skin, platelets, gastrointestinal tissues, and kidney (Hempel et al., 2005). The SULT1A2 gene is located on chromosome 16p11.2-12.1 in close proximity to its related isoform SULT1A1 (Gamage et al., 2006). Besides their classic role in facilitating the detoxification and excretion of their substrates and metabolites, SULT1A1 and SULT1A2 are also known to play a major role in the bioactivation of environmental mutagens and carcinogens, such as hydroxymethyl polycyclic aromatic hydrocarbons, N-hydroxy derivatives of arylamines, allylic alcohols, and heterocyclic amines, leading to mutagenicity and carcinogenesis through the binding of sulfonated metabolites to DNA (Falany, 1997; Weinshilboum et al., 1997; Hempel et al., 2005). Li et al. (2018) recently reported that the expression of liver SULT1A1/2 is highly associated with sex-dependent susceptibility of bladder and liver to the major human bladder carcinogen 4-aminobiphenyl (ABP). Both the parent ABP and its sulfonated metabolites are genotoxic (Chou et al., 1995). In this study, the authors observed that male bladders were more susceptible than female bladders to ABP. This was explained by the increased bladder exposure to ABP in male mice through androgen-dependent suppression of ABP sulfation in the liver, leading to increased bladder delivery of carcinogenic ABP (Li et al., 2018). The male preference in the bladder’s susceptibility to ABP was attenuated by knocking out the Sult1a1 gene. In contrast, female livers were more susceptible than male livers to ABP in mice, which was believed to be due to their higher liver exposure to carcinogens, including APB and its toxic Sult1a1 metabolite, N-sulfate ester-ABP. Further studies revealed that Sult1a1 is positively associated with the increased formation of N-(deoxyguanosin-8-yl)-4-aminobiphenyl, a principal ABP-DNA adduct and the readout of tissue susceptibility to ABP, in hepatic cells. As summarized in Figure 1, androgen renders bladder more exposed to ABP in male mice by suppressing Sult-mediated ABP metabolism in liver, whereas the liver of female mice is more exposed to ABP and its carcinogenic metabolites because of the higher enzymatic activity of liver Sult1a1.
Fig. 1.
Diagram showing the sex- and tissue-specific effect of Sult1a1 on the susceptibility to ABP. In male mice, more ABP was transported into bladder from the liver because of the androgen-dependent suppression of SULT1A1, leading to an increased sensitivity of bladder to ABP-induced carcinogenesis. In female mice, a relatively high expression of SULT1A1 leads to increased sensitivity of the liver to the exposure to ABP and ABP-sulfate. dG-C8-ABP, N-(deoxyguanosin-8-yl)-4-aminobiphenyl.
In humans, it was suggested that the most common polymorphism in SULT1A1 (Arg213His) may have accounted for variations in interindividual susceptibility to hepatocellular carcinoma (HCC) (Boccia et al., 2015; Kim et al., 2015) because SULT1A1 activates environmental mutagens and carcinogens found in well done meat (Bellamri et al., 2018), food processing contaminant (Høie et al., 2016), and dietary flavonoids (Sak and Everaus, 2016). Interestingly, SULT1A1 deactivates carcinogens in cigarette smoke, and a significant interaction between SULT1A1 and smoking was found in a case-control study that included 221 patients with HCC and 290 control subjects (Boccia et al., 2015). Specifically, up to 36% of HCC cases occurred in smokers who carried the Arg213His allele. The high incidence of HCC among Arg213His-carrying smokers was explained by the low catalytic activity of this SULT1A1 variant in detoxifying cigarette smoke carcinogens (Boccia et al., 2015).
SULT1A1 in Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis.
Nonalcoholic fatty liver disease (NAFLD) encompasses a spectrum of diseases ranging from simple steatosis to nonalcoholic steatohepatitis (NASH), which can progress to fibrosis, cirrhosis, and HCC. The pathogeneses of NAFLD and NASH have been extensively studied. However, the roles of SULTs in the initiation and progression of these diseases are insufficiently studied. A hospital-based case control study in an Italian population including 294 NAFLD cases and 359 controls evaluated the effect of the Arg213His SULT1A1 gene polymorphism, which leads to a decreased enzymatic activity of SULT1A1, and the interaction of this polymorphism with lifestyle and dietary habits (Miele et al., 2014). Statistically significant interactions were reported for fruit intake and SULT1A1 gene, as the carriers of Arg213His SULT1A1 gene variant were at higher risk of developing NAFLD when taking high amounts of fruit. The presents of Arg213His SULT1A1 gene variant was also positively associated with grilled meat or fish intake in the development of NAFLD. In addition, many studies found that flavonoids in vegetables and fruits were inhibitors of multiple SULTs (Pai et al., 2001; Huang et al., 2009), which may have affected sensitivity to the development of NAFLD. However, there was no direct evidence linking dietary flavonoid–induced inhibition of SULT1A1 and NAFLD. On the other hand, NAFLD and its progression have a major effect on the expression and activity of multiple SULTs, such as SULT1A1, 1A3, 2A1, and 2B1 (Hardwick et al., 2013; Yalcin et al., 2013). Hardwick et al. (2013) reported an upregulation in protein and enzymatic activity of SULT1A1 in patients with simple steatosis but decreased expression of SULT1A1 in patients with NASH. However, these results are not without controversies. An independent study on normal subjects and patients with steatosis, diabetic cirrhosis, and alcoholic cirrhosis reported a significant decrease of SULT1A1 activity with increasing severity of liver disease from simple steatosis to cirrhosis (Yalcin et al., 2013). A follow-up study from the same group reported that in human liver tissues, sulfation of bisphenol A, an industrial chemical and endocrine disruptor, was substantially lower in livers from subjects with steatosis (23%), diabetic cirrhosis (16%), and cirrhosis (18%) relative to healthy livers (100%), resulting in a higher exposure of bisphenol A in patients with NAFLD (Yalcin et al., 2016). The discrepancies in reported SULT1A1 expression in steatosis might result from the differences in the stages of steatosis and from the gender and age of patients when the liver tissues were collected.
SULT1A1 in Acetaminophen-Induced Liver Injury.
Acetaminophen (APAP), or Tylenol, is one of the most widely used drugs. Although the drug is safe at therapeutic doses, an overdose of APAP remains the leading cause of acute liver failure in the United States. It has been known that human SULT1A1 can catalyze APAP sulfation and facilitate its excretion from the urine. Consistent with their report that the expression and activity of SULT1A1 was increased in steatotic livers and decreased in NASH, Hardwick et al. (2013) showed that the formation of sulfate-APAP was increased in cytosolic fractions of human steatotic liver tissue compared with normal liver but reduced in NASH compared with steatotic liver tissue. A limitation of this study is that the results are largely associations. Future studies are necessary to determine whether SULT1A1 is necessary and sufficient to affect APAP hepatotoxicity.
SULT1A3/4 in Liver Cancers and NAFLD
SULT1A3/4, also known as catecholamine phenol sulfotransferase, play a key role in maintaining the homeostasis of monoamine neurotransmitters, such as dopamine, through sulfation. Unlike SULT1A1/2, which are highly expressed in the hepatocytes, the level of SULT1A3/4 is low in an adult human liver, but they are highly expressed in the fetal liver (Dubaisi et al., 2019) and tumor tissues (Yamamoto et al., 2016). A recent report confirmed that the expression of SULT1A3/4 was increased in tumor tissues compared with adjacent normal tissues of patients with HCC, and the highly expressed SULT1A3/4 in liver tumor tissues was positively associated with its increased enzymatic activity in metabolizing dopamine and elevated metastatic capacity (Zou et al., 2017). Dopamine was a substrate for SULT1A3/4, as well as a regulator responsible for SULT1A3/4 transcriptional activation (Sidharthan et al., 2013). Functional studies showed that the induced expression of SULT1A3/4 in tumor tissues of patients with HCC was negatively correlated with dopamine concentration but positively associated with the epithelial-mesenchymal transition and cancer stem cell acquisition, which ultimately promotes tumor metastasis (Zou et al., 2017).
SULT1A3 activity was reported to be decreased in steatosis, diabetic cirrhosis, and alcoholic cirrhosis liver samples compared with nonfatty control livers (Yalcin et al., 2013), which may have led to reduced sulfation of acetaminophen and opioid drugs (Bairam et al., 2018).
SULT1B1 in Liver Diseases
SULT1B1 plays an important role in the metabolism of drugs, environmental toxins, and endogenous steroids, such as iodothyronines, thyroxine,1-naphthol, and thyroid hormones (Fujita et al., 1997; Wang et al., 1998; Gamage et al., 2006). However, few studies have shown the changes of SULT1B1 expression or activity under different liver disease conditions. According to the Gene Expression Omnibus data base, conflicting observations in SULT1B1 gene expression were reported among different species, the same disease models of the same species under different treatments, or even the same disease models of the same species with the same treatment. As shown in Table 1, there were no consistent changes of SULT1B1 gene expression under HCC or high-fat-diet–induced NASH/NAFLD. Moreover, the lack of study of the roles of SULT1B1 in liver diseases makes it even more challenging to interpret the pathophysiological significance of the altered expression of SULT1B1 in liver diseases.
TABLE 1.
Changes of liver SULT1B1 (Sult1b1) gene expression under different liver disease conditions
| Reporter | Species | Treatment/genotype | Liver disease | Sult1b1 expression |
|---|---|---|---|---|
| GDS3087 | Mouse | Trim 24−/− | HCC | |
| GDS1385 | Rat | Choline-deficient L-amino-acid–defined diet | HCC | |
| GDS5320 | Mouse | Pdgf-c transgenic | HCC | No difference |
| GDS2509 | Mouse | ATP7B−/− | Wilson’s disease | |
| GDS4387 | Human | HBV-associated ALF | No difference | |
| GDS4881 | Human | NAFLD: steatosis | No difference | |
| GDS6248 | Mouse | High fat diet | Liver steatosis | No difference |
| GDS2413 | Mouse | High fat diet | Liver steatosis | |
| GDS4166 | Mouse | High fat diet | Liver steatosis | |
| GDS4817 | Mouse | High fat diet | Liver steatosis | |
| GDS4013 | Mouse | High fat diet | NASH | No difference |
| GDS4013 | Mouse | High fat diet | Pre-NASH | No difference |
| GDS4013 | Mouse | High fat diet | Liver steatosis | No difference |
| GDS4506 | Mouse | ob/ob | Liver steatosis | |
| GDS1354 | Rat | Inhalation of CCl4 | Liver cirrhosis | |
| GDS4271 | Human | Infant biliary atresia: liver | No difference | |
| GDS5526 | Mouse | Bile duct ligation | Liver fibrosis | No difference |
| GDS4389 | Human | Alcoholic hepatitis | ||
| GDS3752 | Mouse | Concanavalin A | Immune-mediated hepatitis |
ALF, acute liver failure; HBV, hepatitis B virus.
SULT1E1
SULT1E1 in Liver Cancer.
Estrogen sulfotransferase (SULT1E1, also known as EST), widely expressed in human tissues such as the liver, lung, adipose tissue, and kidney (Barbosa et al., 2019), is best known for its function in the sulfoconjugation and deactivation of estrogens. This is because sulfonated estrogens fail to bind to the estrogen receptor and thus lose their hormonal activities (Song, 2001). As such, SULT1E1 has long been implicated in female sex hormone–related cancer, such as breast, endometrial, and ovarian cancers (Pasqualini, 2009; Mungenast et al., 2017; Sinreih et al., 2017; Xu et al., 2018). Interestingly, SULT1E1 is also reported to be associated with the occurrence of HCC in rats. In a study on inflammatory liver disease, upregulation of Sult1e1 in diethynitrosamine-treated mouse livers was observed compared with the vehicle-treated livers using real-time reverse-transcription polymerase chain reaction (Lee et al., 2017). However, in the diethynitrosamine-induced rat model of hepatoma, the expression of SULT1E1 was decreased with the onset of hepatomas (Albrethsen et al., 2011). The causal relationship remains to be defined because the expression of SULT1E1 was increased in regenerating liver (Albrethsen et al., 2011). Moreover, the species difference and the human relevance of SULT1E1 in liver cancer need to be clarified.
SULT1E1 in NAFLD and NASH.
Previous study in our laboratory found that loss of Sult1e1/Est in female mice improved metabolic function with improved insulin sensitivity and reduced hepatic steatosis in ob/ob mice lacking Sult1e1 (also known as the obe mice) compared with ob/ob mice (Gao et al., 2012). In an independent study, Sult1e1 was suggested to play a role in sensitizing male mouse to NAFLD/NASH induced by a 20-week feeding of a Western diet high in cholesterol and saturated fat (Matsushita et al., 2017). The authors found that NASH was attenuated in IkbkbΔmye mice with myeloidinhibitor of nuclear factor kappa-B kinase subunit β (IKKβ) IKKβ deficiency in both genders but aggravated in male but not female IkbkbΔhep mice with hepatocyte IKKβ deficiency. Microarray analysis on liver tissues from male and female wild-type (WT) and IkbkbΔhep mice fed high cholesterol and saturated fat diet showed a significant upregulation of Sult1e1 gene in IkbkbΔhep mice compared with WT mice. Among the four groups, the male IkbkbΔhep mice expressed the highest level of Sult1e1 gene, which was associated with decreased plasma estradiol levels. The authors’ mechanism study indicated that the enrichment of liver X receptor (LXR) α to LXRE on the Sult1e1 gene promoter of male IkbkbΔhep mice might be responsible for their heightened sensitivity to NASH. These results suggest hepatocyte IKKβ is protective in males due at least in part to its ability to repress LXR-responsive induction of Sult1e1 (Matsushita et al., 2017).
SULT1E1 in Cholestasis.
Few studies have focused on the role of SULT1E1 in cholestasis, but a recent study suggested that cholestasis-induced farnesoid X receptor (FXR) activation can lead to the suppression of SULT1E1 and hence impede hepatic deactivation of estrogens (Liu et al., 2018). In this study, the authors reported the positive correlation between elevated bile acid levels and estradiol levels in patients with obstructive cholestasis or primary biliary cirrhosis. By using female WT and Fxr−/− mice, the authors went on to show that cholestasis-induced accumulation of estradiol was associated with a downregulation of liver SULT1E1 expression in an FXR-dependent manner. Mechanistic studies using human hepatoma Huh7 cells showed that bile acid activation of FXR repressed SULT1E1 by inhibiting the positive regulation of SULT1E1 by hepatocyte nuclear factor 4α (HNF4α) (Liu et al., 2018).
SULT1E1 in Liver Injury Induced by Sepsis and Ischemia/Reperfusion.
SULT1E1 is relatively thoroughly studied in inflammation-based conditions. Sepsis, resulting from the host’s deleterious systemic inflammatory response to microbial infections, is a major cause of mortality in the intensive care unit. Although sepsis and its associated inflammation are known to decrease the expression and activity of many drug-metabolizing enzymes, we observed a major induction of Sult1e1 and compromised estrogen activity in the liver of mice subjected to the bacterial lipopolysaccharide or cecum ligation and puncture models of sepsis. The inflammatory induction of Sult1e1 gene by sepsis was mediated by nuclear factor κB. Reciprocally, the expression and activity of Sult1e1 can impact the clinical outcome of sepsis. Specifically, ablation of the Sult1e1 gene or pharmacological inhibition of the Sult1e1 enzyme by Triclosan sensitized mice to sepsis-induced death in an estrogen-dependent manner. The increased sepsis-induced lethality in Sult1e1 knockout mice was explained to be due to increased estrogen activity and the resultant attenuated sepsis-induced prosurvival inflammatory response (Chai et al., 2015c).
Hepatic ischemia-reperfusion (I/R) injury is a major cause of liver damage. The pathogenesis of hepatic I/R injury is a dynamic process including the deprivation of blood and oxygen supply during the ischemic phase, followed by their restoration during the reperfusion phase, which is associated with oxidative stress and inflammation. An induction of Sult1e1 in rats subjected to I/R was reported as early as 2006 but without a clearly defined mechanism or understanding of the biologic significance (Svetlov et al., 2006). More recently, we reported a systemic analysis of the effect of liver I/R on the expression of Sult1e1. We showed that the expression of Sult1e1 was markedly induced by I/R in the mouse liver. The ablation of Sult1e1 protected female mice from the injury in an estrogen-dependent manner but heightened liver injury in male mice in an androgen-dependent manner. Further mechanism study established Sult1e1 as a direct transcriptional target of nuclear factor erythroid 2–related factor, a key transcriptional factor responsible for the activation of an array of genes to adapt the cells to hypoxic or oxidative damages. Based on these results, we proposed that inhibition of SULT1E1, at least in females, may represent an effective approach to manage hepatic I/R injury (Guo et al., 2015).
SULT1E1 in Cystic Fibrosis.
Cystic fibrosis (CF), characterized by mutations of both copies of the cystic fibrosis transmembrane receptor (CFTR) gene, is an inherited disorder that causes progressive and eventually fatal damage to the lungs, digestive system, and other organs in the body. Liver is an organ that can be affected by CF. A series of papers published from the laboratory of Charles Falany suggested that the hepatic SULT1E1 may play a role in the progression of liver damage in patients with CF (Li and Falany, 2007; He et al., 2008; Falany et al., 2009). Specifically, elevated hepatic SULT1E1 activity was observed in mouse models of CF (Li and Falany, 2007) and HepG2 cells cocultured with human MMNK-1 cholangiocytes with repressed CFTR (He et al., 2008). The induction of SULT1E1 in CFTR-deficient MMNK-1 cells/HepG2 cells coculture system was found to be dependent on the activation of LXR, and SULT1E1 induction led to alterations in the expression of estrogen responsive genes, including insulin-like growth factor 1 (IGF-1), glutathione S-transferase P1 (GST-P1), and carbonic anhydrase II, due to decreased estradiol levels. These results suggest that the induction of hepatic SULT1E1 in patients with CF may have facilitated the development of CF liver disease (Falany et al., 2009).
SULT2
SULT2A1
SULT2A1 in Cholestasis.
Cholestasis is an accumulation of bile acids in the liver as a result of increased bile acid production in the liver and/or insufficient detoxication and elimination of bile acids from the liver. SULT2A1, highly expressed in the liver and adrenal gland, mainly catalyzes the sulfation of hydroxysteroids, including bile acids (Radominska et al., 1990), hydroxysteroid dehydroepiandrosterone (Otterness et al., 1992), and androgens. SULT2A1 is reported to play an important role in the detoxification of bile acids and thereby preventing cholestasis via its enzymatic activity in bile acid sulfation. The expression of SULT2A1 is transcriptionally regulated by several nuclear receptors, such as pregnane X receptor (PXR; Sonoda et al., 2002), constitutive androstane receptor (Saini et al., 2004), LXRα (Uppal et al., 2007; Ou et al., 2014), HNF4α (Fang et al., 2007), and FXR (Song et al., 2001). The activation of SULT2A1 by PXR, constitutive androstane receptor, and LXRα was believed to play critical roles in the anticholestatic activity of these receptors in rodent models of cholestasis (Sonoda et al., 2002; Saini et al., 2004; Ou et al., 2014). In humans, it was reported that liver SULT2A1 expression is decreased in human obstructive cholestasis due to gallstone biliary obstruction (Chai et al., 2015b). In a more recent clinical study, Wunsch et al. (2015) showed that liver SULT2A1 expression is decreased in patients with primary sclerosing cholangitis, which was proposed to be due to microRNA-378a-5p–mediated inhibition of the PXR/SULT2A1 axis. Table 2 summarizes several studies suggesting that activation of SULT2A1 expression through drug treatment protects against the development of cholestasis.
TABLE 2.
SULT2A1 expression in cholestasis
| Compounds | Disease models | Species | Expression of Sult2a1 | Mediators | Clinical outcome | References |
|---|---|---|---|---|---|---|
| Yangonin | Estrogen-induced cholestasis | Mouse | FXR | Attenuation | Gao et al., 2018; Dong et al., 2019 | |
| Auraptene | 17α-Ethinylestradiol–induced cholestasis | Mouse | FXR | Attenuation | Wang et al., 2019 | |
| Tamoxifen | Cholestasis | Human | / | Enhancement | Bansal and Lau, 2019 | |
| Calculus bovis sativus | Estrogen-induced cholestasis | Rat | FXR | Attenuation | Xiang et al., 2019 | |
| N-3 polyunsaturated fatty acids | / | Human cell line | / | Induce BA detoxification | Cieślak et al., 2018 | |
| Corilagin | ANIT-induced cholestasis | Rat | FXR | Attenuation | Yang et al., 2018 | |
| Doxorubicin | Cholic acid-induced cholestasis | Mouse | p53 | Attenuation | Chen et al., 2017 | |
| Geniposide | ANIT-induced cholestasis | Rat | FXR | Attenuation | Wang et al., 2017 | |
| Alisol B 23-acetate | Estrogen-induced cholestasis | Mouse | FXR | Attenuation | Meng et al., 2015a | |
| Alisol B 23-acetate | ANIT-induced cholestasis | Mouse | FXR | Attenuation | Meng et al., 2015b | |
| Oleanolic acid | Bile duct ligation–induced cholestasis | Rat | / | Attenuation | Chai et al., 2015a | |
| Oleanolic acid | Lithocholic acid–induced cholestasis | Mouse | ERR2 | Attenuation | Chen et al., 2014 |
ANIT, alpha-naphthylisothiocyanate; BA, bile acid; ERR2, estrogen receptor–related receptor 2.
SULT2A1 in Other Liver Diseases.
Downregulations of SULT2A1 gene were found in human HCC (Huang et al., 2005) and alcoholic liver disease (Yang et al., 2019b), but not in NASH (Suga et al., 2019). An increased liver Sult2a1 gene expression was observed in choline-deficient, L-amino-acid–defined, high-fat-diet–induced mouse model of NASH (Suga et al., 2019).
SULT2B1b
SULT2B1b in Hepatocellular Carcinoma.
The cholesterol sulfotransferase SULT2B1b catalyzes the sulfation of cholesterol to synthesize cholesterol sulfate. Yang et al. (2013) found that SULT2B1b expression promotes the proliferation of HCC cells, which may have contributed to the progression of HCC. Specifically, the expression of SULT2B1b was found to be higher in HCC tumor tissues than their adjacent normal tissues. Moreover, overexpression of SULT2B1b promoted the growth of the mouse HCC Hepa1-6 cells, whereas knockdown of SULT2B1b inhibited the cell growth with induced cell-cycle arrest and apoptosis via upregulating the expression of fatty acid synthase, downregulating the expression of CyclinB1, BCL2, and MYC in vitro and in vivo. A follow-up mechanistic study from the same group reported that SULT2B1b promotes the proliferation of HCC cells via the activation of the β-catenin/matrix metalloproteinase 7 pathway in hepatocytes and therefore enhances epithelial-mesenchymal transition (Yang et al., 2019a).
SULT2B1b in NAFLD and Metabolic Liver Disease.
SULT2B1b is also reported to play an important role in NAFLD. Overexpression of SULT2B1b decreased serum and hepatic lipids in mouse models of NAFLD via the suppression of LXR/sterol regulatory element binding protein (SREBP) 1c signaling pathway (Bai et al., 2012). In their study, the authors found SULT2B1b overexpression, combined with administration of the LXR agonist 25-hydroxycholesterol, significantly increased the formation of sulfated metabolite of 25-hydroxycholesterol in the liver tissue and decreased serum and hepatic lipid levels, including triglycerides, total cholesterol, free cholesterol, and free fatty acids, as compared with the control group both in WT and Ldlr−/− mice. SULT2B1b may have inhibited steatosis by sulfonation and deactivation of oxysterols, the endogenous agonists for the lipogenic nuclear receptor LXR. Indeed, gene expression analysis showed that overexpression of SULT2B1b was accompanied by the reduced expression of LXR target lipogenic genes, such as SREBP-1, SREBP-2, acetyl-CoA carboxylase-1, and fatty acid synthase (Bai et al., 2012). The inhibition of NAFLD was also observed in transgenic mice that overexpress SULT2B1b in the liver (Shi et al., 2014).
In addition to inhibiting lipogenesis, SULT2B1b and its metabolic byproduct cholesterol sulfate exhibit major activity in inhibiting hepatic gluconeogenesis and relieving metabolic liver disease (Shi et al., 2014; Bi et al., 2018). Initially, we reported that SULT2B1b can inhibit hepatic gluconeogenesis by suppressing the gluconeogenic activity of HNF4α (Shi et al., 2014). In this study, we found that the treatment with cholesterol sulfate or transgenic overexpression of SULT2B1b in the liver inhibited hepatic gluconeogenesis and attenuated metabolic abnormalities in both the high-fat-diet–induced obesity and ob/ob mice. At the mechanistic level, treatment with cholesterol sulfate or overexpression of SULT2B1b inhibited gluconeogenesis via the suppression of acetyl-CoA synthetase expression, leading to decreased acetylation and nuclear exclusion of HNF4α. In a subsequent study, we reported that SULT2B1b can be transcriptionally and positively regulated by HNF4α (Bi et al., 2018), an observation that led to our hypothesis that the transactivation of SULT2B1b by HNF4α represents a negative feedback to limit the gluconeogenic activity of HNF4α. Consistent with our hypothesis we showed that downregulation of Sult2B1b promoted the gluconeogenic activity of HNF4α as a result of decreased expression of the HNF4α deacetylase Sirt1 and increased HNF4α acetylation. Sult2b1b expression was also induced by HNF4α upon fasting. Ablation of Sult2B1b in mice led to increased gluconeogenic gene expression and an elevated fasting glucose level, suggesting that SULT2B1b plays a restrictive role in HNF4α-mediated fasting responsive gluconeogenesis. In the same study, we also designed and synthesized thiocholesterol, a hydrolysis-resistant derivative of cholesterol sulfate, which showed more superior activity than the native cholesterol sulfate in inhibiting gluconeogenesis and improving insulin sensitivity in high-fat-diet–induced diabetic mice. Based on these results, we conclude that the HNF4α-SULT2B1b–cholesterol sulfate axis represents a key endogenous mechanism to prevent uncontrolled gluconeogenesis.
SULT2B1b in APAP-Induced Liver Injury.
In a recent study, we uncovered an unexpected role for SULT2B1b in APAP-induced liver injury. Hepatic overexpression of SULT2b1b enhanced the sensitivity of mice to APAP-induced liver injury, whereas ablation of the Sult2B1b in mice conferred resistance to the APAP hepatotoxicity (An et al., 2019). This is a somewhat surprising result considering that sulfation is generally considered to be a metabolic pathway that detoxifies APAP. Consistent with our previous finding that SULT2B1b is a transcriptional target of HNF4α, Hnf4α overexpression also sensitized mice to APAP-induced hepatotoxicity in a Sult2b1b-dependent manner, indicating that the HNF4α-SULT2B1b axis plays a unique role in APAP-induced hepatotoxicity and that SULT2B1b induction might be a risk factor for APAP toxicity (An et al., 2019).
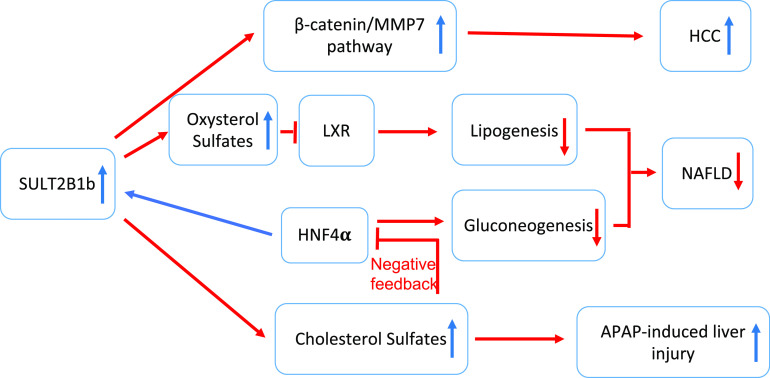
As summarized in Figure 2, SULT2B1b plays important roles in the development of liver cancer, NAFLD, and sensitivity to APAP-induced liver injury.
Fig. 2.
Summary of the role of SULT2B1B in liver diseases. Overexpression of SULT2B1b promotes the progression of HCC via the activation of the β-catenin/matrix metalloproteinase 7 (MMP7) pathway in hepatocytes. Overexpression of SULT2B1b increases the sulfonation and deactivation of oxysterols and thereby dampens the lipogenic function of LXR and decreases the serum and hepatic lipids. In addition to inhibiting lipogenesis, SULT2B1b and its metabolic byproduct cholesterol sulfate exhibit major activity in inhibiting hepatic gluconeogenesis and relieving metabolic liver disease by suppressing the gluconeogenic activity of HNF4α, a transcription factor that positively regulates SULT2B1b gene expression. The induction of SULT2B1b by HNF4α represents a negative feedback to limit the gluconeogenic activity of HNF4α. Overexpression of SULT2B1b sensitized mice to APAP-induced liver injury, and overexpression of Hnf4α increases the sensitivity to APAP-induced hepatotoxicity in a Sult2b1b-dependent manner.
Summary and Perspectives
Over the past 20 years or so, significant advances have been achieved in our understanding of the roles of SULTs in multiple liver diseases, including liver cancers, nonalcoholic liver diseases, and drug-induced liver injury. Accumulating evidence shows altered expression and/or regulation of SULTs in liver diseases can reciprocally alter clinical consequences of liver diseases. Sex-, tissue-, and development-specific regulations of SULTs, such as SULT1A1/2 and SULT1E1, appeared to be important factors affecting the pathogenesis of liver diseases. In most cases, the effect of the SULTs on liver diseases can be explained by the sulfonation of xenobiotics and endobiotics, which has a major impact on the chemical and functional homeostasis of these chemicals.
Despite the progress in our understanding of the role of SULTs in liver diseases, there are a number of remaining questions and challenges: 1) the identification of previously unknown pathophysiologic functions of SULT isoforms and understanding the molecular mechanisms underlying the roles of SULT beyond regulating the sulfation and deactivation of a variety of endogenous and exogenous molecules; 2) exploring SULTs and SULT-mediated signaling pathways as therapeutic targets for disease conditions such as NAFLD/NASH, sepsis- or ischemia-reperfusion–induced liver injury, drug-induced liver injury, and even cancers; 3) investigating the species specificity of the SULTs’ function, as the majority of the in vivo and mechanistic studies have been focused on rodents; human studies are necessary to determine whether the rodent results are translatable to humans; and 4) the humanization of cytochrome P450 enzymes, such as CYP3A4 (Granvil et al., 2003; Cheung et al., 2006) and 2D6 (Corchero et al., 2001) have been reported, but the humanization of SULTs is a largely unexplored territory.
Abbreviations
- ABP
4-aminobiphenyl
- APAP
acetaminophen
- CF
cystic fibrosis
- CFTR
cystic fibrosis transmembrane receptor
- FXR
farnesoid X receptor
- IGF-1
insulin-like growth factor 1
- HCC
hepatocellular carcinoma
- HNF4α
hepatocyte nuclear factor 4α
- IKKβ
inhibitor of nuclear factor kappa-B kinase subunit β
- I/R
ischemia-reperfusion
- LXR
liver X receptor
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- PAPS
phosphoadenosine-5′-phosphosulfate
- PXR
pregnane X receptor
- SREBP
sterol regulatory element binding protein
- SULT
sulfotransferase
- WT
wild type
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Y. Xie, W. Xie.
Footnotes
Our original work described in this review article was supported in part by National Institutes of Health [Grants DK117370 and ES030429] (to W.X.). W.X. was supported in part by the Joseph Koslow Endowed Professorship from the University of Pittsburgh School of Pharmacy.
The data contained in this manuscript were presented in part from the doctoral dissertation of Y.X. (http://d-scholarship.pitt.edu/38357/).
References
- Albrethsen J, Miller LM, Novikoff PM, Angeletti RH. (2011) Gel-based proteomics of liver cancer progression in rat. Biochim Biophys Acta 1814:1367–1376. [DOI] [PubMed] [Google Scholar]
- Ambadapadi S, Wang PL, Palii SP, James MO. (2017) Celecoxib affects estrogen sulfonation catalyzed by several human hepatic sulfotransferases, but does not stimulate 17-sulfonation in rat liver. J Steroid Biochem Mol Biol 172:46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y, Wang P, Xu P, Tung HC, Xie Y, Kirisci L, Xu M, Ren S, Tian X, Ma X, et al. (2019) An unexpected role of cholesterol sulfotransferase and its regulation in sensitizing mice to acetaminophen-induced liver injury. Mol Pharmacol 95:597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Q, Zhang X, Xu L, Kakiyama G, Heuman D, Sanyal A, Pandak WM, Yin L, Xie W, Ren S. (2012) Oxysterol sulfation by cytosolic sulfotransferase suppresses liver X receptor/sterol regulatory element binding protein-1c signaling pathway and reduces serum and hepatic lipids in mouse models of nonalcoholic fatty liver disease. Metabolism 61:836–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairam AF, Rasool MI, Alherz FA, Abunnaja MS, El Daibani AA, Kurogi K, Liu MC. (2018) Effects of human SULT1A3/SULT1A4 genetic polymorphisms on the sulfation of acetaminophen and opioid drugs by the cytosolic sulfotransferase SULT1A3. Arch Biochem Biophys 648:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S, Lau AJ. (2019) Inhibition of human sulfotransferase 2A1-catalyzed sulfonation of lithocholic acid, glycolithocholic acid, and taurolithocholic acid by selective estrogen receptor modulators and various analogs and metabolites. J Pharmacol Exp Ther 369:389–405. [DOI] [PubMed] [Google Scholar]
- Barbosa ACS, Feng Y, Yu C, Huang M, Xie W. (2019) Estrogen sulfotransferase in the metabolism of estrogenic drugs and in the pathogenesis of diseases. Expert Opin Drug Metab Toxicol 15:329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann E. (1876) Ueber α-Kresylschwefelsäure. Eur J Inorg Chem 9:1389–1392. [Google Scholar]
- Bellamri M, Xiao S, Murugan P, Weight CJ, Turesky RJ. (2018) Metabolic activation of the cooked meat carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in human prostate. Toxicol Sci 163:543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y, Shi X, Zhu J, Guan X, Garbacz WG, Huang Y, Gao L, Yan J, Xu M, Ren S, et al. (2018) Regulation of cholesterol sulfotransferase SULT2B1b by hepatocyte nuclear factor 4α constitutes a negative feedback control of hepatic gluconeogenesis. Mol Cell Biol 38:e00654-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerregaard-Olesen C, Bossi R, Bech BH, Bonefeld-Jørgensen EC. (2015) Extraction of perfluorinated alkyl acids from human serum for determination of the combined xenoestrogenic transactivity: a method development. Chemosphere 129:232–238. [DOI] [PubMed] [Google Scholar]
- Boccia S, Miele L, Panic N, Turati F, Arzani D, Cefalo C, Amore R, Bulajic M, Pompili M, Rapaccini G, et al. (2015) The effect of CYP, GST, and SULT polymorphisms and their interaction with smoking on the risk of hepatocellular carcinoma. BioMed Res Int 2015:179867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J, Du X, Chen S, Feng X, Cheng Y, Zhang L, Gao Y, Li S, He X, Wang R, et al. (2015a) Oral administration of oleanolic acid, isolated from Swertia mussotii Franch, attenuates liver injury, inflammation, and cholestasis in bile duct-ligated rats. Int J Clin Exp Med 8:1691–1702. [PMC free article] [PubMed] [Google Scholar]
- Chai J, Feng X, Zhang L, Chen S, Cheng Y, He X, Yang Y, He Y, Wang H, Wang R, et al. (2015b) Hepatic expression of detoxification enzymes is decreased in human obstructive cholestasis due to gallstone biliary obstruction. PLoS One 10:e0120055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai X, Guo Y, Jiang M, Hu B, Li Z, Fan J, Deng M, Billiar TR, Kucera HR, Gaikwad NW, et al. (2015c) Oestrogen sulfotransferase ablation sensitizes mice to sepsis. Nat Commun 6:7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Li D, Chen Y, Sun J, Fu K, Guan L, Zhang H, Jiang Y, Li X, Zeng X, et al. (2017) p53-mediated regulation of bile acid disposition attenuates cholic acid-induced cholestasis in mice. Br J Pharmacol 174:4345–4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Zeng H, Wang Y, Fan X, Xu C, Deng R, Zhou X, Bi H, Huang M. (2014) Low dose of oleanolic acid protects against lithocholic acid-induced cholestasis in mice: potential involvement of nuclear factor-E2-related factor 2-mediated upregulation of multidrug resistance-associated proteins. Drug Metab Dispos 42:844–852. [DOI] [PubMed] [Google Scholar]
- Cheung C, Yu AM, Chen CS, Krausz KW, Byrd LG, Feigenbaum L, Edwards RJ, Waxman DJ, Gonzalez FJ. (2006) Growth hormone determines sexual dimorphism of hepatic cytochrome P450 3A4 expression in transgenic mice. J Pharmacol Exp Ther 316:1328–1334. [DOI] [PubMed] [Google Scholar]
- Chou HC, Lang NP, Kadlubar FF. (1995) Metabolic activation of N-hydroxy arylamines and N-hydroxy heterocyclic amines by human sulfotransferase(s). Cancer Res 55:525–529. [PubMed] [Google Scholar]
- Cieślak A, Trottier J, Verreault M, Milkiewicz P, Vohl MC, Barbier O. (2018) N-3 polyunsaturated fatty acids stimulate bile acid detoxification in human cell models. Can J Gastroenterol Hepatol 2018:6031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corchero J, Granvil CP, Akiyama TE, Hayhurst GP, Pimprale S, Feigenbaum L, Idle JR, Gonzalez FJ. (2001) The CYP2D6 humanized mouse: effect of the human CYP2D6 transgene and HNF4alpha on the disposition of debrisoquine in the mouse. Mol Pharmacol 60:1260–1267. [DOI] [PubMed] [Google Scholar]
- Dong R, Wang J, Gao X, Wang C, Liu K, Wu J, Liu Z, Sun H, Ma X, Meng Q. (2019) Yangonin protects against estrogen-induced cholestasis in a farnesoid X receptor-dependent manner. Eur J Pharmacol 857:172461. [DOI] [PubMed] [Google Scholar]
- Dooley TP, Haldeman-Cahill R, Joiner J, Wilborn TW. (2000) Expression profiling of human sulfotransferase and sulfatase gene superfamilies in epithelial tissues and cultured cells. Biochem Biophys Res Commun 277:236–245. [DOI] [PubMed] [Google Scholar]
- Dubaisi S, Caruso JA, Gaedigk R, Vyhlidal CA, Smith PC, Hines RN, Kocarek TA, Runge-Morris M. (2019) Developmental expression of the cytosolic sulfotransferases in human liver. Drug Metab Dispos 47:592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falany CN. (1991) Molecular enzymology of human liver cytosolic sulfotransferases. Trends Pharmacol Sci 12:255–259. [DOI] [PubMed] [Google Scholar]
- Falany CN. (1997) Enzymology of human cytosolic sulfotransferases. FASEB J 11:206–216. [DOI] [PubMed] [Google Scholar]
- Falany CN, He D, Li L, Falany JL, Wilborn TW, Kocarek TA, Runge-Morris M. (2009) Regulation of hepatic sulfotransferase (SULT) 1E1 expression and effects on estrogenic activity in cystic fibrosis (CF). J Steroid Biochem Mol Biol 114:113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang HL, Strom SC, Ellis E, Duanmu Z, Fu J, Duniec-Dmuchowski Z, Falany CN, Falany JL, Kocarek TA, Runge-Morris M. (2007) Positive and negative regulation of human hepatic hydroxysteroid sulfotransferase (SULT2A1) gene transcription by rifampicin: roles of hepatocyte nuclear factor 4alpha and pregnane X receptor. J Pharmacol Exp Ther 323:586–598. [DOI] [PubMed] [Google Scholar]
- Fang JL, Wu Y, Gamboa da Costa G, Chen S, Chitranshi P, Beland FA. (2016) Human sulfotransferases enhance the cytotoxicity of tolvaptan. Toxicol Sci 150:27–39. [DOI] [PubMed] [Google Scholar]
- Fujita K, Nagata K, Ozawa S, Sasano H, Yamazoe Y. (1997) Molecular cloning and characterization of rat ST1B1 and human ST1B2 cDNAs, encoding thyroid hormone sulfotransferases. J Biochem 122:1052–1061. [DOI] [PubMed] [Google Scholar]
- Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME. (2006) Human sulfotransferases and their role in chemical metabolism. Toxicol Sci 90:5–22. [DOI] [PubMed] [Google Scholar]
- Gao J, He J, Shi X, Stefanovic-Racic M, Xu M, O’Doherty RM, Garcia-Ocana A, Xie W. (2012) Sex-specific effect of estrogen sulfotransferase on mouse models of type 2 diabetes. Diabetes 61:1543–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Fu T, Wang C, Ning C, Liu K, Liu Z, Sun H, Ma X, Huo X, Yang X, et al. (2018) Yangonin protects against cholestasis and hepatotoxity via activation of farnesoid X receptor in vivo and in vitro. Toxicol Appl Pharmacol 348:105–116. [DOI] [PubMed] [Google Scholar]
- Glatt H, Boeing H, Engelke CE, Ma L, Kuhlow A, Pabel U, Pomplun D, Teubner W, Meinl W. (2001) Human cytosolic sulphotransferases: genetics, characteristics, toxicological aspects. Mutat Res 482:27–40. [DOI] [PubMed] [Google Scholar]
- Granvil CP, Yu AM, Elizondo G, Akiyama TE, Cheung C, Feigenbaum L, Krausz KW, Gonzalez FJ. (2003) Expression of the human CYP3A4 gene in the small intestine of transgenic mice: in vitro metabolism and pharmacokinetics of midazolam. Drug Metab Dispos 31:548–558. [DOI] [PubMed] [Google Scholar]
- Guo Y, Hu B, Huang H, Tsung A, Gaikwad NW, Xu M, Jiang M, Ren S, Fan J, Billiar TR, et al. (2015) Estrogen sulfotransferase is an oxidative stress-responsive gene that gender-specifically affects liver ischemia/reperfusion injury. J Biol Chem 290:14754–14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick RN, Ferreira DW, More VR, Lake AD, Lu Z, Manautou JE, Slitt AL, Cherrington NJ. (2013) Altered UDP-glucuronosyltransferase and sulfotransferase expression and function during progressive stages of human nonalcoholic fatty liver disease. Drug Metab Dispos 41:554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, Wilborn TW, Falany JL, Li L, Falany CN. (2008) Repression of CFTR activity in human MMNK-1 cholangiocytes induces sulfotransferase 1E1 expression in co-cultured HepG2 hepatocytes. Biochim Biophys Acta 1783:2391–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel N, Negishi M, McManus ME. (2005) Human SULT1A genes: cloning and activity assays of the SULT1A promoters. Methods Enzymol 400:147–165. [DOI] [PubMed] [Google Scholar]
- Herrero J, Muffato M, Beal K, Fitzgerald S, Gordon L, Pignatelli M, Vilella AJ, Searle SM, Amode R, Brent S, et al. (2016) Ensembl comparative genomics resources [published correction appears in Database (Oxford) (2016) 2016:baw053]. Database (Oxford) 2016:bav096.26896847 [Google Scholar]
- Høie AH, Monien BH, Glatt H, Hjertholm H, Husøy T. (2016) DNA adducts induced by food mutagen PhIP in a mouse model expressing human sulfotransferases 1A1 and 1A2. Toxicol Lett 248:34–38. [DOI] [PubMed] [Google Scholar]
- Huang C, Chen Y, Zhou T, Chen G. (2009) Sulfation of dietary flavonoids by human sulfotransferases. Xenobiotica 39:312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Bathena SP, Tong J, Roth M, Hagenbuch B, Alnouti Y. (2010) Kinetic analysis of bile acid sulfation by stably expressed human sulfotransferase 2A1 (SULT2A1). Xenobiotica 40:184–194. [DOI] [PubMed] [Google Scholar]
- Huang LR, Coughtrie MW, Hsu HC. (2005) Down-regulation of dehydroepiandrosterone sulfotransferase gene in human hepatocellular carcinoma. Mol Cell Endocrinol 231:87–94. [DOI] [PubMed] [Google Scholar]
- Jancova P, Anzenbacher P, Anzenbacherova E. (2010) Phase II drug metabolizing enzymes. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 154:103–116. [DOI] [PubMed] [Google Scholar]
- Kauffman FC. (2004) Sulfonation in pharmacology and toxicology. Drug Metab Rev 36:823–843. [DOI] [PubMed] [Google Scholar]
- Kim IW, Han N, Kim MG, Kim T, Oh JM. (2015) Copy number variability analysis of pharmacogenes in patients with lymphoma, leukemia, hepatocellular, and lung carcinoma using the Cancer Genome Atlas data. Pharmacogenet Genomics 25:1–7. [DOI] [PubMed] [Google Scholar]
- Krattinger R, Boström A, Lee SML, Thasler WE, Schiöth HB, Kullak-Ublick GA, Mwinyi J. (2016) Chenodeoxycholic acid significantly impacts the expression of miRNAs and genes involved in lipid, bile acid and drug metabolism in human hepatocytes. Life Sci 156:47–56. [DOI] [PubMed] [Google Scholar]
- Lee SR, Lee SY, Kim SY, Ryu SY, Park BK, Hong EJ. (2017) Hydroxylation and sulfation of sex steroid hormones in inflammatory liver. J Biomed Res 31:437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Falany CN. (2007) Elevated hepatic SULT1E1 activity in mouse models of cystic fibrosis alters the regulation of estrogen responsive proteins. J Cyst Fibros 6:23–30. [DOI] [PubMed] [Google Scholar]
- Li Y, Chen Z, Paonessa JD, Meinl W, Bhattacharya A, Glatt H, Vouros P, Zhang Y. (2018) Strong impact of sulfotransferases on DNA adduct formation by 4-aminobiphenyl in bladder and liver in mice. Cancer Med 7:5604–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Xue R, Yang C, Gu J, Chen S, Zhang S. (2018) Cholestasis-induced bile acid elevates estrogen level via farnesoid X receptor-mediated suppression of the estrogen sulfotransferase SULT1E1. J Biol Chem 293:12759–12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita N, Hassanein MT, Martinez-Clemente M, Lazaro R, French SW, Xie W, Lai K, Karin M, Tsukamoto H. (2017) Gender difference in NASH susceptibility: roles of hepatocyte Ikkβ and Sult1e1. PLoS One 12:e0181052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Chen X, Wang C, Liu Q, Sun H, Sun P, Huo X, Liu Z, Yao J, Liu K. (2015a) Protective effects of alisol B 23-acetate via farnesoid X receptor-mediated regulation of transporters and enzymes in estrogen-induced cholestatic liver injury in mice. Pharm Res 32:3688–3698. [DOI] [PubMed] [Google Scholar]
- Meng Q, Chen XL, Wang CY, Liu Q, Sun HJ, Sun PY, Huo XK, Liu ZH, Yao JH, Liu KX. (2015b) Alisol B 23-acetate protects against ANIT-induced hepatotoxity and cholestasis, due to FXR-mediated regulation of transporters and enzymes involved in bile acid homeostasis. Toxicol Appl Pharmacol 283:178–186. [DOI] [PubMed] [Google Scholar]
- Miele L, Dall’armi V, Cefalo C, Nedovic B, Arzani D, Amore R, Rapaccini G, Gasbarrini A, Ricciardi W, Grieco A, et al. (2014) A case-control study on the effect of metabolic gene polymorphisms, nutrition, and their interaction on the risk of non-alcoholic fatty liver disease. Genes Nutr 9:383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JW, Gilligan LC, Idkowiak J, Arlt W, Foster PA. (2015) The regulation of steroid action by sulfation and desulfation. Endocr Rev 36:526–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JW, Idkowiak J, Gesteira TF, Vallet C, Hardman R, van den Boom J, Dhir V, Knauer SK, Rosta E, Arlt W. (2018) Human DHEA sulfation requires direct interaction between PAPS synthase 2 and DHEA sulfotransferase SULT2A1. J Biol Chem 293:9724–9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungenast F, Aust S, Vergote I, Vanderstichele A, Sehouli J, Braicu E, Mahner S, Castillo-Tong DC, Zeillinger R, Thalhammer T. (2017) Clinical significance of the estrogen-modifying enzymes steroid sulfatase and estrogen sulfotransferase in epithelial ovarian cancer. Oncol Lett 13:4047–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi M, Pedersen LG, Petrotchenko E, Shevtsov S, Gorokhov A, Kakuta Y, Pedersen LC. (2001) Structure and function of sulfotransferases. Arch Biochem Biophys 390:149–157. [DOI] [PubMed] [Google Scholar]
- Otterness DM, Wieben ED, Wood TC, Watson WG, Madden BJ, McCormick DJ, Weinshilboum RM. (1992) Human liver dehydroepiandrosterone sulfotransferase: molecular cloning and expression of cDNA. Mol Pharmacol 41:865–872. [PubMed] [Google Scholar]
- Ou Z, Jiang M, Hu B, Huang Y, Xu M, Ren S, Li S, Liu S, Xie W, Huang M. (2014) Transcriptional regulation of human hydroxysteroid sulfotransferase SULT2A1 by LXRα. Drug Metab Dispos 42:1684–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai TG, Suiko M, Sakakibara Y, Liu MC. (2001) Sulfation of flavonoids and other phenolic dietary compounds by the human cytosolic sulfotransferases. Biochem Biophys Res Commun 285:1175–1179. [DOI] [PubMed] [Google Scholar]
- Pasqualini JR. (2009) Estrogen sulfotransferases in breast and endometrial cancers. Ann N Y Acad Sci 1155:88–98. [DOI] [PubMed] [Google Scholar]
- Radominska A, Comer KA, Zimniak P, Falany J, Iscan M, Falany CN. (1990) Human liver steroid sulphotransferase sulphates bile acids. Biochem J 272:597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, Weinshilboum RM. (1997) Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem Biophys Res Commun 239:298–304. [DOI] [PubMed] [Google Scholar]
- Raftogianis RB, Wood TC, Weinshilboum RM. (1999) Human phenol sulfotransferases SULT1A2 and SULT1A1: genetic polymorphisms, allozyme properties, and human liver genotype-phenotype correlations. Biochem Pharmacol 58:605–616. [DOI] [PubMed] [Google Scholar]
- Reinen J, Vermeulen NP. (2015) Biotransformation of endocrine disrupting compounds by selected phase I and phase II enzymes--formation of estrogenic and chemically reactive metabolites by cytochromes P450 and sulfotransferases. Curr Med Chem 22:500–527. [DOI] [PubMed] [Google Scholar]
- Saeki Y, Sakakibara Y, Araki Y, Yanagisawa K, Suiko M, Nakajima H, Liu MC. (1998) Molecular cloning, expression, and characterization of a novel mouse liver SULT1B1 sulfotransferase. J Biochem 124:55–64. [DOI] [PubMed] [Google Scholar]
- Saini SP, Sonoda J, Xu L, Toma D, Uppal H, Mu Y, Ren S, Moore DD, Evans RM, Xie W. (2004) A novel constitutive androstane receptor-mediated and CYP3A-independent pathway of bile acid detoxification. Mol Pharmacol 65:292–300. [DOI] [PubMed] [Google Scholar]
- Sak K, Everaus H. (2016) Sulfotransferase 1A1 as a biomarker for susceptibility to carcinogenesis: from molecular genetics to the role of dietary flavonoids. Curr Drug Metab 17:528–541. [DOI] [PubMed] [Google Scholar]
- Shi X, Cheng Q, Xu L, Yan J, Jiang M, He J, Xu M, Stefanovic-Racic M, Sipula I, O’Doherty RM, et al. (2014) Cholesterol sulfate and cholesterol sulfotransferase inhibit gluconeogenesis by targeting hepatocyte nuclear factor 4α. Mol Cell Biol 34:485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidharthan NP, Minchin RF, Butcher NJ. (2013) Cytosolic sulfotransferase 1A3 is induced by dopamine and protects neuronal cells from dopamine toxicity: role of D1 receptor-N-methyl-D-aspartate receptor coupling. J Biol Chem 288:34364–34374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinreih M, Knific T, Anko M, Hevir N, Vouk K, Jerin A, Frković Grazio S, Rižner TL. (2017) The significance of the sulfatase pathway for local estrogen formation in endometrial cancer. Front Pharmacol 8:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CS, Echchgadda I, Baek BS, Ahn SC, Oh T, Roy AK, Chatterjee B. (2001) Dehydroepiandrosterone sulfotransferase gene induction by bile acid activated farnesoid X receptor. J Biol Chem 276:42549–42556. [DOI] [PubMed] [Google Scholar]
- Song WC. (2001) Biochemistry and reproductive endocrinology of estrogen sulfotransferase. Ann N Y Acad Sci 948:43–50. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Xie W, Rosenfeld JM, Barwick JL, Guzelian PS, Evans RM. (2002) Regulation of a xenobiotic sulfonation cascade by nuclear pregnane X receptor (PXR). Proc Natl Acad Sci USA 99:13801–13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strott CA. (2002) Sulfonation and molecular action. Endocr Rev 23:703–732. [DOI] [PubMed] [Google Scholar]
- Suga T, Yamaguchi H, Ogura J, Shoji S, Maekawa M, Mano N. (2019) Altered bile acid composition and disposition in a mouse model of non-alcoholic steatohepatitis. Toxicol Appl Pharmacol 379:114664. [DOI] [PubMed] [Google Scholar]
- Svetlov SI, Xiang Y, Oli MW, Foley DP, Huang G, Hayes RL, Ottens AK, Wang KK. (2006) Identification and preliminary validation of novel biomarkers of acute hepatic ischaemia/reperfusion injury using dual-platform proteomic/degradomic approaches. Biomarkers 11:355–369. [DOI] [PubMed] [Google Scholar]
- Uppal H, Saini SP, Moschetta A, Mu Y, Zhou J, Gong H, Zhai Y, Ren S, Michalopoulos GK, Mangelsdorf DJ, et al. (2007) Activation of LXRs prevents bile acid toxicity and cholestasis in female mice. Hepatology 45:422–432. [DOI] [PubMed] [Google Scholar]
- Wang D, Li Q, Favis R, Jadwin A, Chung H, Fu DJ, Savitz A, Gopal S, Cohen N. (2014) SULT4A1 haplotype: conflicting results on its role as a biomarker of antipsychotic response. Pharmacogenomics 15:1557–1564. [DOI] [PubMed] [Google Scholar]
- Wang J, Falany JL, Falany CN. (1998) Expression and characterization of a novel thyroid hormone-sulfating form of cytosolic sulfotransferase from human liver. Mol Pharmacol 53:274–282. [DOI] [PubMed] [Google Scholar]
- Wang J, Fu T, Dong R, Wang C, Liu K, Sun H, Huo X, Ma X, Yang X, Meng Q. (2019) Hepatoprotection of auraptene from the peels of citrus fruits against 17α-ethinylestradiol-induced cholestasis in mice by activating farnesoid X receptor. Food Funct 10:3839–3850. [DOI] [PubMed] [Google Scholar]
- Wang L, Wu G, Wu F, Jiang N, Lin Y. (2017) Geniposide attenuates ANIT-induced cholestasis through regulation of transporters and enzymes involved in bile acids homeostasis in rats. J Ethnopharmacol 196:178–185. [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM, Otterness DM, Aksoy IA, Wood TC, Her C, Raftogianis RB. (1997) Sulfation and sulfotransferases 1: sulfotransferase molecular biology: cDNAs and genes. FASEB J 11:3–14. [PubMed] [Google Scholar]
- Wunsch E, Klak M, Wasik U, Milkiewicz M, Blatkiewicz M, Urasinska E, Barbier O, Bielicki D, Bogdanos DP, Elias E, et al. (2015) Liver expression of sulphotransferase 2A1 enzyme is impaired in patients with primary sclerosing cholangitis: lack of the response to enhanced expression of PXR. J Immunol Res 2015:571353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang D, Yang J, Liu Y, He W, Zhang S, Li X, Zhang C, Liu D. (2019) Calculus bovis sativus improves bile acid homeostasis via farnesoid X receptor-mediated signaling in rats with estrogen-induced cholestasis. Front Pharmacol 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Yan TM, Chen JM, Li XY, Zou J, Zhu LJ, Lu LL, Wang Y, Zhou FY, Liu ZQ, et al. (2017) LC-MS/MS quantification of sulfotransferases is better than conventional immunogenic methods in determining human liver SULT activities: implication in precision medicine. Sci Rep 7:3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Lin X, Xu J, Jing H, Qin Y, Li Y. (2018) SULT1E1 inhibits cell proliferation and invasion by activating PPARγ in breast cancer. J Cancer 9:1078–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin EB, Kulkarni SR, Slitt AL, King R. (2016) Bisphenol A sulfonation is impaired in metabolic and liver disease. Toxicol Appl Pharmacol 292:75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin EB, More V, Neira KL, Lu ZJ, Cherrington NJ, Slitt AL, King RS. (2013) Downregulation of sulfotransferase expression and activity in diseased human livers. Drug Metab Dispos 41:1642–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Kurogi K, Schiefer IT, Liu MY, Sakakibara Y, Suiko M, Liu MC. (2016) Human cytosolic sulfotransferase SULT1A3 mediates the sulfation of dextrorphan. Biol Pharm Bull 39:1432–1436. [DOI] [PubMed] [Google Scholar]
- Yang F, Wang Y, Li G, Xue J, Chen ZL, Jin F, Luo L, Zhou X, Ma Q, Cai X, et al. (2018) Effects of corilagin on alleviating cholestasis via farnesoid X receptor-associated pathways in vitro and in vivo. Br J Pharmacol 175:810–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Du X, Sun L, Zhao X, Zhu J, Li G, Tian J, Li X, Wang Z. (2019a) SULT2B1b promotes epithelial-mesenchymal transition through activation of the β-catenin/MMP7 pathway in hepatocytes. Biochem Biophys Res Commun 510:495–500. [DOI] [PubMed] [Google Scholar]
- Yang X, Xu Y, Guo F, Ning Y, Zhi X, Yin L, Li X. (2013) Hydroxysteroid sulfotransferase SULT2B1b promotes hepatocellular carcinoma cells proliferation in vitro and in vivo. PLoS One 8:e60853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Kusumanchi P, Ross RA, Heathers L, Chandler K, Oshodi A, Thoudam T, Li F, Wang L, Liangpunsakul S. (2019b) Serum metabolomic profiling identifies key metabolic signatures associated with pathogenesis of alcoholic liver disease in humans. Hepatol Commun 3:542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yetti H, Naito H, Yuan Y, Jia X, Hayashi Y, Tamada H, Kitamori K, Ikeda K, Yamori Y, Nakajima T. (2018) Bile acid detoxifying enzymes limit susceptibility to liver fibrosis in female SHRSP5/Dmcr rats fed with a high-fat-cholesterol diet. PLoS One 13:e0192863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Varlamova O, Vargas FM, Falany CN, Leyh TS. (1998) Sulfuryl transfer: the catalytic mechanism of human estrogen sulfotransferase [published correction appears in J Biol Chem (1998) 273:17296]. J Biol Chem 273:10888–10892. [DOI] [PubMed] [Google Scholar]
- Zou J, Li H, Huang Q, Liu X, Qi X, Wang Y, Lu L, Liu Z. (2017) Dopamine-induced SULT1A3/4 promotes EMT and cancer stemness in hepatocellular carcinoma. Tumour Biol 39:1010428317719272. [DOI] [PubMed] [Google Scholar]