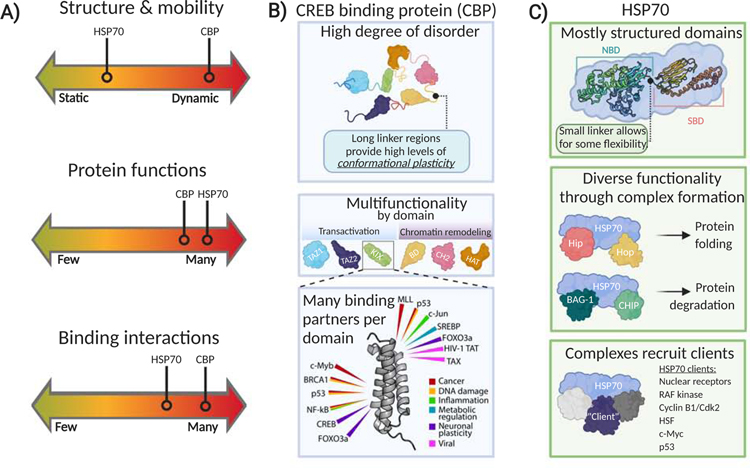
Figure 1. Key attributes of dynamic proteins with representative examples.
(A) While no single definition can be used to identify dynamic proteins, useful metrics for successful classification include: 1) the structure and mobility of the protein and/or the individual domains; 2) the number of functional roles the protein can fulfill; and 3) the number binding interactions the protein can participate in. Dynamic proteins tend to be multifunctional, participating in many pathways and performing multiple cellular roles, either through the presence of multiple domains or by the formation of dynamic transient complexes that vary in function. The functional multiplicity can in part be attributed to the number of binding partners the protein engages with, which oftentimes leads to conformational changes either within a single domain or domain rearrangement as an entire protein to elicit variable functional effects. (B) The master coactivator CREB binding protein (CBP) analyzed as a representative dynamic protein. CBP is highly disordered, with more unstructured and dynamic regions than structured domains (Dyson and Wright, 2016). The protein has high conformational plasticity, as its domains are connected by long unstructured linkers. The individual domains within CBP allow it to perform multiple functions. For example, CBP has four activator binding domains, KIX, TAZ1, TAZ2, and IBiD, that interact with transcriptional activators to regulate gene expression. It also contains a histone acetyltransferase domain (HAT) and bromodomain (BD) that it utilizes for perform chromatin remodeling functions (Breen and Mapp, 2018). Each of these individual domains has its own network of interactions, summing up to 100s of potential PPIs made by CBP. The KIX domain is shown here as an example – it interacts with a suite of 15+ activator proteins, all which can be implicated in a variety of diseases (Mapp et al., 2015). (C) The chaperone HSP70 analyzed as a representative dynamic protein. HSP70 contains much more secondary structure than CBP. It is comprised of two main domains: the nucleotide binding domain (NBD) and substrate binding domain (SBD). These two domains are connected by a small dynamic linker (Fernández-Fernández and Valpuesta, 2018). Substrate binding can induce domain rearrangement to elicit different functional effects. HSP70 itself functions as an ATPase, however it is able to perform many diverse cellular functions by forming different multiprotein complexes. For example, Hsp70 in complex with cochaperones Hip and Hop acts as a protein folding complex, while Hsp70 in complex with BAG-1 and CHIP functions as a degradation complex (Muller et al., 2013). Finally, Hsp70 complexes also interact with many other proteins. Many regulatory proteins are known to be controlled via transient association with Hsp70. Thus, Hsp70 is a potential point of therapeutic intervention for conditions such as cancer, autoimmune and neurodegenerative diseases (A. Assimon et al., 2013).