Figure 7.
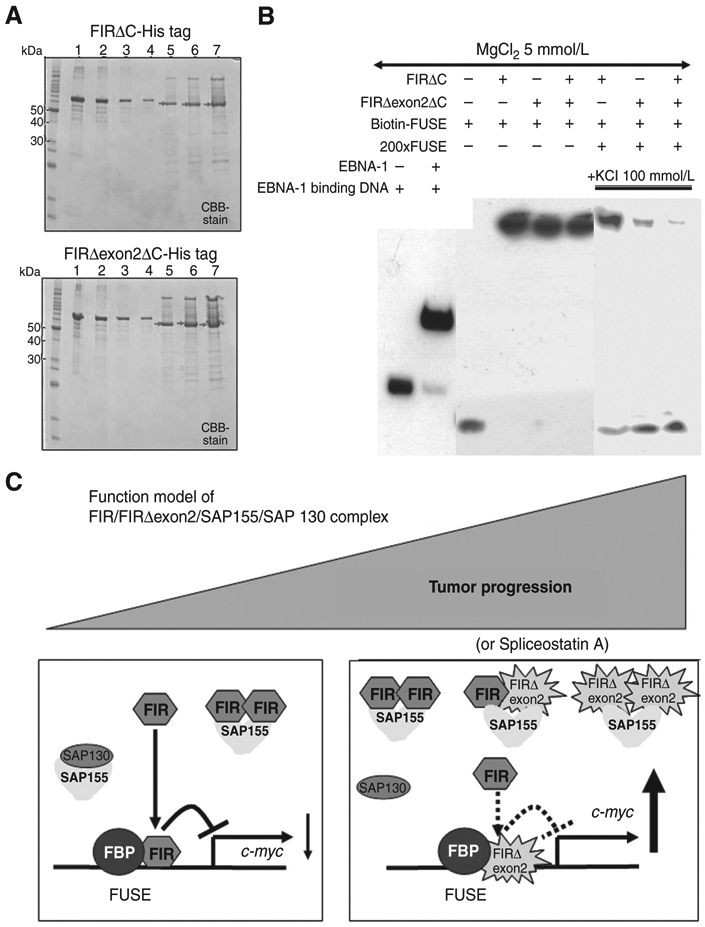
FIRΔexon2 potentially interferes with FIR to bind to FUSE along with tumor progression. A, C-terminal 95 amino acids deleted FIR- or FIRΔexon2-His tag protein, FIRΔC- or FIRΔexon2ΔC-His tag or with FIR-RRM1 and RRM2 (amino acids 103–297, exons 6–9), was prepared, purified, and was analyzed by SDS-PAGE. After affinity purification by HisTrapHP, recovered sample was dialyzed against 50 mmol/L Tris-HCl (pH9.0). Lane 1; 1,000 ng of bovine serum albumin (BSA), lane 2; 500 ng of BSA, lane 3; 250 ng of BSA, lane 4; 125 ng of BSA, lane 5; 2 μL lane 6; 4 μL, lane 7; 8 μL of purified FIR- (top) or FIRΔexon2-His tag (bottom) proteins. Concentration of FIRΔexon2-His tag proteins was estimated by band intensity using with BSA as a standard. B, EMSA revealed that FIRΔexon2ΔC interferes with FIRΔC to bind to FUSE. C, functional model of FIR/FIRΔexon2/SAP155/SAP130 complex along with tumor progression. FIR suppresses c-myc gene transcription and SAP155 engages in alternative splicing in noncancer cells, whereas potential FIR, FIRΔexon2, and SAP155 complex disturbed authentic FIR and SAP155 function simultaneously in cancer cells.
